The Third Revolution

The Third Revolution
If we were to ask someone randomly what the most important discoveries in biology have been in the course of its history, the answers we would most likely receive are (1) The Darwinian theory of evolution by natural selection and (2) the discovery of the structure of DNA. Indeed, these are both perfectly correct answers. Yet there has been a third crucial revolution in biology, which has occurred only in the last 30 years or so, and is just as tremendous and important as the two mentioned above, but is perhaps overlooked outside the community of specialists because it is not the result of a single monumental event but is rather an amalgam of discoveries, improved techniques and scientific reasoning. This is the third revolution of biology.
The achievement I am talking about is precisely this: A complete formulation of the properties of biological organisms in terms entirely of chemistry.
Although the idea of a “vital essence” that distinguished biological organisms from inorganic objects has been rejected by the scientific community for over 100 years, it has only been in the last 30 years that we have been able to describe exactly how chemistry and biology relate. There is no longer a “gap” between the two. The monumentality of this achievement has only been possible with the rise of highly advanced analytical techniques and technology, such as Protein NMR spectroscopy, X-Ray crystallography, Polymerase Chain Reactions, and the rise of tremendous computing power and databanking employed by biologists in determining the relationships between DNA sequences hundreds of millions of base pairs long, and between homologous proteins with tremendously complex and still not fully understood 3D structures. In short, it has only been possible in the last 20-30 years.
That is precisely the topic of this essay. During this process we will be able to answer a very fundamental question in a very precise way: What is biological life, and how is it related to chemistry?
In order to grasp the nature of this relationship, the reader must be familiar with the fundamental principles of organic chemistry and a significant portion of thermodynamic chemistry as well, not to mention atomic chemistry and the differing natures of types of chemical bonds. Furthermore, a topic such as this requires a very high degree of precision. We cannot use simplified and pared down versions of actual occurrences. We cannot use vague notions like “DNA holds the information to create an organism” and expect this to actually aid our understanding of this subject. Notions like this are far too vague to be useful here. We will be jettisoning a large amount of vague terminology here in our attempts to elucidate exactly what occurs. Whenever we do use concepts that seem vague at first such as “chemical pathway” or “information” we will define these in exact terms in due course.
I should warn the reader that the material which follows is very hard. Some of it is introductory but some of it, especially near the end, would not be out of place in a 2nd or 3rd year undergraduate lecture. Nonetheless, as we shall see, and as I often stress, the difficulty of molecular biology lies not in the underlying principles, but in the combinatorics of what we are studying. No matter how complicated a process we might examine seems, the reader should (and, hopefully, will) appreciate that the unifying principles are identical.
It may seem like an absolutely monumental task to elucidate the exact nature of the relationship between chemistry and biology, but while discovering the information was a monumental task, actually pinning it down in words, now that it is known, is that not difficult. Despite the hundreds of thousands of complex biological pathways uncovered and the vast number of complex chemical interactions that occur in biological systems, there are only a handful of underlying principles that govern all of them, and all of these principles refer directly to three distinct types of linear, unbranched polymers, namely DNA, RNA and protein. It may be best to elucidate exactly what these principles are before we work toward what they actually mean. I have narrowed it down to six:
1. The ability of nucleic acids to act as a template for its own synthesis by polymerization and the synthesis of related nucleic acids by virtue of complementary hydrogen bonding and hence the ability of DNA to act as a template for RNA
2. The ability of nucleic to act as templates for the synthesis of proteins by virtue of triplet sequences of nucleic acids which correspond to particular amino acids
3. The ability of proteins to, by means of non-covalent interactions between carboxyl, amine and side chain groupings, to fold into distinct 3D shapes giving them the totality of their individual properties
4. The ability of proteins to form hydrogen bonded contacts with the outer face nucleotides on DNA polymers and hence for the protein molecules to alter the activity of regions of DNA (the contact of which can induce allosteric changes in the binding proteins)
5. The ability of RNA molecules to form ribonucleoprotein complexes by virtue of hydrogen bonded contacts with matching surfaces of polypeptides
6. The ability of polypeptides to form protein complexes by means of matching surfaces via hydrogen bonding and hence for allosteric mechanical changes to be induced in the binding proteins or small ligands resulting in a change of structure and function,
These six properties of linear unbranched polymers which are involved directly or indirectly in everything interesting that occurs in the cells constitute the totality of the link between chemistry and biology. Note that these six properties are not totally different. In fact, they are basically the same, and can be summed up like this: All biological occurrences can be characterized entirely in terms of the properties of three large organic polymers (RNA, DNA and Protein) built from a set of monomers such that the interactions between different monomers give rise to complex polymeric properties characterizing all biological phenomena.
At the atomic level, there is nothing special about a biological organism. There is no essence stamped or imbued on them. The carbon atoms in them are no more special than the carbon atoms in the graphite in your pencil, the nitrogen atoms in them no more special than those in the atmosphere. It is the chemical properties of the large organic molecules which make up organisms which sets biology apart. And those properties in turn, are purely the result of physical forces between the atoms inside the polymers.
Now that this relationship has been elucidated exactly, the hard part comes. We must find out exactly what this means. The expert reader will already know exactly what they mean and appreciate them. Such readers are asked to bear with me until we move to discussing more complex and interesting biological phenomena. First, we must have a little primer in organic chemistry and atomic chemistry. It is important to stress that this is merely prerequisite knowledge and has been known since long before the relationship between chemistry and biology had been formalized.
In a biological organism we would expect a vast variety of different atoms to be participating in the bulk of the chemical reactions and pathways. But there are not. In fact, the truly vast majority of everything that constitutes you and any other biological organism is composed of just four elements; Carbon, hydrogen, oxygen and nitrogen. With that list I have accounted for essentially the entire mass of a biological organism. RNA, DNA and other polymers such as glycogen, starch and lipids are composed entirely of these atoms. Proteins contain sulfur as well due to methionine and cysteine, and these sulfur groups prove important in protein folding, and ions of manganate, magnesium, potassium, sodium and chloride prove to be crucial ligands in reactions and conductors of electricity in neurons, but the actual three types of macromolecules giving rise to all interesting properties of biology are composed entirely of carbon, hydrogen, oxygen, nitrogen and sulfur.
How is this possible? The answer lies in the discipline of organic chemistry. Organic chemistry is also called carbon chemistry and with good reason. Carbon lies at the very heart of this whole discussion. Despite being 70% water by mass, we are called “carbon based life forms” again with good reason.
Carbon and Four covalent bonds
The particular property of carbon making it so crucial is its ability to form four covalent bonds. To understand precisely what this means and why this is important requires us to delve into atomic and bonding chemistry. Since this discussion is taking a highly reductionist approach it makes perfect sense that we should start from the ground up, considering properties of single atoms, before discussing the properties of macromolecules, and in turn, biological structures.
Covalent bonding is the strongest type of bonding that can occur between two atoms (a bond between two atoms being defined as a chemical bond). There are 5 types of chemical bond and fortunately they are all based on exactly the same principle: The distribution of valence electrons between atoms in a bond. Chemistry is sometimes called the study of valence electrons and with good reason: Because that’s what it is. All chemical interactions are characterized in terms of changes in distribution of electrons in atoms. (The concept of a unifying principle is cropping up quite a lot, as we can see! This is a good thing. It means that our scientific understanding is highly unified and has excellent explanatory power, which is exactly what we want). The five types of bonding mentioned are covalent, ionic, hydrogen bonding, van der Waals forces and dipole-dipole interaction. Strictly speaking, ionic and covalent bonds should not be considered two distinct types of bonds since they are just positions on a sliding scale of electronegativity, the degree to which a particular atom attracts electrons.
The details of different electronegativities of atoms are not important (we cannot go too far backward in terms of reductionism or we will just end up veering off the topic of the essay. This is a good place to start from.). The principle we must take away here is that the sharing of electrons between two atoms characterizes both ionic and covalent bonds. If there is a very large difference in electronegativity between the two atoms in question, then the bond formed is said to be highly polar because one molecule becomes negatively charged (the one which has a much greater tendency to hold the electron) and the other positively. The result is an ionic compound, which is held together by purely electrostatic forces. Ionic compounds will disassociate in water to become their individual ions (not individual atoms since they are now charged as one has gained and the other lost electrons, and atoms by definition are uncharged). As a consequence, ionic compounds never occur in biology because everything takes place in the fluid of the cell.
If the electronegativity difference between two atoms is sufficiently small, then the sharing of electrons is more even and hence the bond is said to be covalent. Covalent bonds are the strongest bonds by several orders of magnitude and (this is crucial) they define individual molecules. Thus, if two or more atomic species are all covalently bonded with each other then they have formed a molecule, but if two molecules hydrogen bond with each other they have not formed one larger molecule. If the electronegativity difference between two atoms is sufficiently small as to not be ionic, but sufficiently large that the shared electrons tend significantly to one side of the bond, then one side will become slightly charged negatively and other positively. This is called a polar covalent bond and lies at the very heart of biochemistry.

Thus, we are now in a position to talk about carbon’s formation of four covalent bonds. This fact allows for the construction of vast molecules with carbons chained together and other species attached in the other spaces which each carbon atom can accommodate (again, the details of why this occurs is not important, but if you are interested it is because of the octet rule).

This is what sets organic chemistry apart from inorganic chemistry. In inorganic chemistry we do not encounter very large molecules. Most commonly, we encounter small covalent molecules, and metals (which don’t form covalent molecules at all, but instead form metallic bonds, which are totally different). But in organic chemistry, we have the opportunity to work with single molecules which are huge. It is crucial to understand that a DNA double helix of a single chromosome is only consistent of two molecules (one for each helix). Each helix is one very, very, very large straight polymer held together by covalent bonds containing, on average (in a human chromosome) 8,000,000,000 atoms. These are the largest known single molecules.
The existence of polar covalent bonds allows for the weak interaction between the polar covalent bonds of different molecules to form significantly weaker non-covalent interactions. These non-covalent interactions occurred in all six underlying chemical properties mentioned at the start, so obviously they are completely central. There are three sorts that concern us.
Hydrogen bonding:
This is the strongest type of non-covalent interaction. It occurs when a highly polar covalent group containing hydrogen on a molecule is oriented with a a lone pair on another atom in another covalent group. A lone pair is simply a pair of electrons which do not participate in a covalent bond. Between small molecules, hydrogen bonding occurs between different molecules, but in large straight molecules which can twist and fold, hydrogen bonding can occur between different segments of the same molecule as well as between two large molecules. The latter interactions are at the core of the six properties listed earlier. Hydrogen bonds are weak relative to covalent bonds but compared to dipole-dipole and especially VDW forces are very strong. They are, for example, the reason that ethanol (which can form hydrogen bonds) boils at 352K but ethanal (which can only form dipole-dipole interactions) boils at 293K.

Dipole-Dipole Forces
As we have seen, molecules containing atoms of different electronegativity will have polar bonds. This results in one of the atoms being negative and the other positive. These molecules will have an attractive force between them called dipole-dipole forces. These are much weaker than hydrogen bonds. This has a lesser effect on physical properties than hydrogen bonds because it is significantly weaker.
Since we have just discussed dipole-dipole forces it seems reasonable to include the following absolutely crucial point here which we absolutely must remember for the rest of the essay. Over the course of the essay a vast number of distinct types of complex chemical interactions between different types of molecules will be detailed. We will see that biological systems are defined by these chemical processes, and that these processes occur within a distinct physically bounded chemical system called cells. The crucial point about cells is that all biological processes within cells occur within an aqueous environment, in other words they occur within the medium of water. Thus, contained within the physical boundary that defines a single cell are the series of complex chemical reactions which we shall soon see define biological life and that these interactions occur within a fluid that fills the system called the cytoplasm.
Van der Waals Forces
Non-polar covalently bonded molecules experience a weaker intermolecular force between their molecules called the van der Waals forces. This results from the temporary flickering dipole that occurs when the molecules get close enough to each other for the electrons in one orbital to induce a very short lived dipole in the other. This is by far the weakest type of intermolecular forces. Molecules with only this type of intermolecular force between them have very low melting points. Since VDW forces depend on the interaction between electrons in two particles, the greater the molar mass, the greater the VDW interaction between the electrons. This is absolutely crucial in biology. Despite the weakness of VDW forces, they become noticeable in the interaction between two or more very large polymers.
There is a general principle underlying all three types of non-covalent interaction. Firstly, they are all attractive in nature. They are not repulsive. As a consequence, the strength of bonding between two large molecules is characterized by the number of non-covalent interactions formed. This depends on the orientation of particular groups and the number and polarity of these groups present, all of which determine the fold and structure of a protein, as we shall see.
Organic chemistry and functional groups
This understanding of bonding allows us to move into organic chemistry. The good thing about organic chemistry is that it is so systematic and modular; in fact it is arguably the most systematically formed branch of any discipline of science. Carbon forms the basis of all organic compounds. More precisely chains of carbon atoms form the basis of organic compounds and functional groups with different properties are attached to these carbon chains to create different molecules with different properties. The most common bond that occurs in organic chemistry is the C-H bond. In many ways it is a “default bond” because it is so stable and is in fact not very interesting since it is completely non-polar but it does provide the backbone of organic compounds. A chain composed entirely of carbons bound to hydrogens is called an alkyl chain and many organic compounds are based on alkyl chains with functional groups attached.
A set of the most important and by far the most common functional groups is shown below.

We should note properties of these organic compounds immediately. Bonds like N-H and C=O and C-O and O-H are polar whereas bonds like C-H are not. We shall appreciate the importance of this more when we come to the monomers which make up the three biological polymers already discussed.
In general, an organic compound has physical and chemical properties defined by three things. One is the chain length. The second is the number and type of functional groups attached and the third is the structural isomerism. Because all of the interesting polymeric molecules we are discussing are linear polymers and because biological systems are stereo-selective, we will not be discussing isomerism here. But the fact that a molecule can be characterized by chain length and functional group gives rise to the fundamental notion in organic chemistry of a homologous series. A homologous series is simply a family of organic compounds which differ only by the length of their alkyl chain.
Shown above are the first four of the homologous series of alkanes which are given as R-H where R is an alkyl chain of arbitrary length. In general, there are many sorts of alkyl chains and they naturally give rise to a highly systematic nomenclature, but they are in fact, not the central organic molecules of study of molecular biology. Biological polymers, as we shall see, are not comprised of homologous series but instead of repeating units of monomers which are drawn from distinct families of small organic molecules.
Polymers and Monomers
Distinct families of small organic molecules characterized by particular functional groups can give rise to families of biochemicals which constitute the basis of biological polymers. In order to understand this the idea of a monomer and a polymer must be elucidated. In organic chemistry, the ability of carbon to form four covalent bonds allows for the creation of long chained molecules formed from repeating units. The individual repeating units are called monomers. There are only two sorts of monomers actually concerning us here. They are nucleotides and amino acids. The former constitute the monomers of DNA and RNA, the latter, proteins. A general amino acid takes the following form:

Let us identify the functional groups present here. We can see a carboxyl group, an amine group and a side chain. A side chain is not a particular functional group but it is what defines the particular amino acids. The presence of the carboxylic acid group and the amine group is what defines an amino acid, and the side chain is what defines a particular amino acid, it is what makes it unique. There are 20 biological amino acids and they are shown below. It is the particular side chains that give each amino acid its properties.

We should note from the table above that we can classify the amino acids on the basis of their chemical groups. A side chain can be non-polar or it can be polar. A polar amino acid can either be charged (negatively or positively) or uncharged. We shall see that sequences of differently polarized and charged amino acids in proteins is of primary importance in determining the final structure of a 3D protein.
The polymerization of these amino acids occurs between the carboxylic acid and amine groups to form an amide linkage by chemists and a peptide bond by biologists. The formation of amide linkages in sequence is what gives rise to polypeptides. The sequence of amino acids is therefore what defines a particular polymer. As the amide linkages form, the side chain groups point out to the side. The order of amino acids in a polypeptide is what determines the order of different side chains and hence interaction between side chains and hence the 3D structure which forms. Despite that proteins are constitute of linear chains of amino acids, they do not in general have linear straight line physical appearance. Proteins fold into 3D conformations discussed in more detail below.

This shows the linking of two amino acids to form a dipeptide. The same principle is repeated thousands of times in biological polypeptides in which amino acids link in linear sequences thousands of amino acids long. The diagram below shows four amino acids linking in sequence to form tetrapeptide. The same principle applies to polpeptides, extending for thousands of amino acids instead of four.

We come now to the polymers of DNA and RNA. Fortunately, these are vastly simpler than protein. The biological function of a protein is determined by its final 3D structure after it has folded. But DNA does not work like this, its usefulness and function comes not from its 3D structure and it does not fold into highly complex structures*, but instead it is determined purely from a particular chemical property of its monomers which allows its sequence to serve as a template. We shall soon see what this means. DNA stands for deoxyribonucleic acid. It, like RNA (which stands for ribonucleic acid) is a nucleic acid polymer. The monomers which constitute this polymer are nucleotides. As analogous to proteins, there are different types of nucleotides and their sequence in a nucleic acid is what determines the properties of a sequence of DNA. However, whereas there are 20 amino acids occurring in biology**, there are only four nucleotides which occur in DNA and likewise only four*** nucleotides occurring in RNA (three of which are the same as those occurring in DNA). EAs a monomer a nucleotide is composed of three distinct groups, a phosphate group, a sugar group and finally a base. There is a clear analogy with amino acids, whereas all amino acids have carboxyl and amine groups and are distinguished as particular amino acids via their side chains, all nucleotides possess a sugar-phosphate grouping. It is their base which determines them. The general form of a nucleotide is shown below:

The four bases of DNA are shown below:
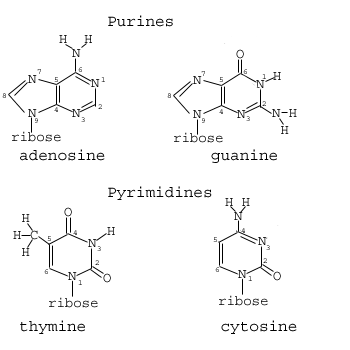
Note that the bases can be divided into two distinct classes: Those with single and those with double rings. Those with single rings are called pyrimidine rings. Those with double rings are purines which are composed of adjacent pyrimidine and imidazole rings. Something similar can be said for RNA, which differs from DNA in only one of four bases and with the addition of an extra oxygen grouping on the sugar component of the nucleotide:

Whereas amino acids link via their peptide linkages, nucleotides form straight chain nucleic acids via phosphodiester linkages. We can see a clear analogue here again. As nucleic acids form, the bases are aligned along the sides of the polymer just as the side chains of the amino acids point out and away from the peptide bond. These phosphodiester linkages are shown below:

The above picture shows two nucleotides linking to form a dinucleotide. Just as amino acids, the same principle applies for thousands (or, actually, in this case, millions) of nucleotides to form polynucleotides. DNA and RNA polymers are both polynucleotides. Short sequences of four linked nucleotides of RNA and DNA monomers are shown below. Clearly this principle is exactly the same as the polymerization of amino acids:

*DNA does fold but it does so for the purpose of compaction, since having a single molecule of DNA stretch end to end in the nucleus would be analogous to packing a tennis ball with enough string to circumnavigate the Earth several times. As we shall see, this compaction does have an important role to play in the mechanisms of cells.
**technically there are 21 due to selenocysteine, but this is only inserted through special modification and is uncommon
***technically there are 5 due to inosine, but this is only inserted due to special modification and is uncommon
Thus in general there is an underlying principle to all three major polymers being discussed. They are all straight chain polymers comprised from a defined set of unit monomers the sequence of which in the polymer determining its properties.
Having discussed the chemical structure of RNA, DNA and protein in terms of their monomers, we can now turn to how the sequences of these monomers in the polymers they form give rise to the properties of the polymers which are responsible for biological life. We shall start with protein folding.
Protein Folding
So important is protein folding and so difficult to understand that it is essentially a discipline on its own, a discipline which has only been possible with the use of supercomputers, since humans are not capable of distinguishing between very precise structural similarities occurring in homologous proteins and sorting through the vast amount of actual information that we have about the relationship between structural motifs and sequences.
Early experiments using in vitro analysis made a crucial discovery which would found this discipline. When, for example, an enzyme is placed in a solution in vitro it will continue to perform its function of catalysis. In other words, it will maintain its folded 3D structure which determines its catalytic properties. When treated with something like sodium hydroxide the protein will unfold entirely and will no longer work. Once the sodium hydroxide has been neutralized the protein will continue to catalyze again.
Experiments like this demonstrate a key property of polypeptides. They can spontaneously fold into their shape, which determines their properties. They do not, in general, need help in vivo to fold. Thus all the information determining the confirmation and hence the properties of a particular protein is contained within its amino acid sequence.
In principle, this means that we should be able to take a protein whose sequence of amino acids is known, but whose structure is not, feed the sequence into a supercomputer and be told the structure. This is called the protein folding problem. At present, although we do have a very large amount of information about particular sequences and structural motifs, we have neither the sufficient computing power nor the knowledge of protein structure to be able to do this. Protein structure must be determined via other means such as X-Ray crystallography.
There are a certain number of general principles of protein folding which should be expounded upon here because they are instructive in terms of understanding how sequences of amino acids give rise to 3D structures.
Arguably the most important principle is the tendency of hydrophobic stretches of amino acids to cluster on the interior of the protein so as to exclude water. We have seen that the side chains of 8 amino acids are non-polar. The interaction between non-polar molecules and water gives rise to a hydrophobic force. The idea of a hydrophobic force was not considered above in discussing non-covalent interactions because it is a fictional force. It is not a consequence of electrostatic interactions between molecules but rather is due to the fact that non polar molecules mixing with water is thermodynamically unfavorable. Water is an extremely polar molecule. It is so polar, in fact, that if one were to take a strip of plastic cloth, rub it vigorously with a cloth so as to induce a static charge distribution, and then place it near a thin stream of running water, they could get the stream of water to bend toward the strip.
In the presence of a highly non-polar molecule, the water molecules will arrange themselves so as to minimize contact with the non-polar groupings. Non-polar groupings in turn, being hydrophobic will act so as to minimize contact with the water molecules by forming contacts with each other. This is shown below:

This is part of a general principle. Non-polar molecules are not miscible with polar molecules. The consequence of this for protein molecules is that stretches of hydrophobic segments will fold up so as to contact with each other in the interior of a protein. If a protein is malformed then hydrophobic segments will present themselves on the exterior face of the protein. This is extremely dangerous for it will cause other malformed proteins to form cluster contacts with their malformed segments to exclude water. As a consequence, the malformed proteins can aggregate and precipitate out of solution. This is exactly what happens to someone suffering from Alzheimer’s disease.
Hydrophobic forces are an example of contact between side chains causing a particular structural feature of a protein, but contacts between the amine and carboxyl groups within the protein are important too. The N-H bond is very, very polar, with the H atom on the bond being very positive. The C=O grouping on the carboxyl component of the amine linkage is also very polar, with the oxygen possessing a lone pair. As a consequence, the N-H groupings will tend to form hydrogen bonds with nearby C=O bonds. This occurs spontaneously during the process of protein folding and gives rise to two key structural motifs. These are alpha helices and Beta-sheets, and these motifs are ubiquitous throughout proteomics. Both of these are shown below:


The ubiquity of these motifs means that in a schematic representation of a protein they will always be shown. Their importance lies in the way that these highly repeated motifs can create particular structures which give rise to the highly diverse group of properties of particular proteins. This is discussed later. Because of their ubiquity, schematic diagrams depict proteins usually entirely in terms of them, as shown below:

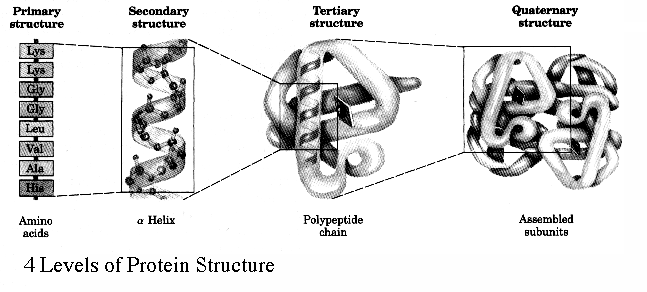
We have seen a few examples of motifs and general rules which proteins conform to when folding. It is therefore time to introduce some terminology. Proteins in general fold very quickly and spontaneously. In fact, they fold while they are still being made. The sequence of amino acids which constitute the protein is simply called the primary structure of the protein. These sequences of amino acids are responsible for the motifs of Beta-sheets and alpha helices that form during the process of protein folding. These motifs are the secondary structure. Once all the secondary motifs have arranged themselves into the final 3D conformation of the polypeptide then the tertiary structure has been reached. At this point, the object in question is a completely folded polypeptide, but completed proteins in the cell are never found as single polypeptides, but large scale complexes of multiple polypeptides attached to each other in non-covalently held together proteins, with each polypeptide being referred to as a protein subunit. Once multiple tertiary subunits have come together to form a completed protein, the result is the quaternary structure.
The general principle we should take away is that the secondary motifs are the result of the interactions between the backbone carboxyl and amine groupings and are held together by hydrogen bonds. More complex interactions that define individual chains as they fold up will involve interactions between side chains which result in the formation of complex regions which give the protein its properties.
DNA and RNA as templates of their own polymerization
No doubt the amount of information presented above seems exhausting. Unfortunately, that was by and large merely prerequisite knowledge, the bare necessity to begin to understand the relationship between biology and chemistry. The time has now come to discuss the discovery of Watson and Crick, which was the start of the third revolution.
I have said before that the important property of DNA is its ability to guide its own synthesis via templated polymerization by means of complementary hydrogen bonds. It is now time to articulate exactly what this means. We have seen that molecules of DNA are unbranched polymers composed of 4 types of nucleotides. Recall from above, that these four nucleotides are defined by their different bases, which are adenine, thymine, guanine and cytosine henceforth denoted A,T,G and C respectively. Also recall from above that two of these bases are pyrimidines and two are purines.
Here, then is the crucial realization of Watson and Crick: The two purines and two pyrimidines that form the monomers of DNA form complementary purine-pyrimidine pairs. By “complementary pairs” we mean they recognize their complement by virtue of hydrogen bonding. A nucleotide monomer in DNA can only form strong hydrogen bonds with its respective complement. The complement of the pyrimidine T is the purine A and vice-versa. The complement of the pyrimidine C is the purine G and vice versa.

This complementary base pairing thus allows for templated polymerization. If we have a polymeric sequence of nucleotides, we can employ this polymeric sequence to create the complement polymer, because nucleotide monomers will slot into place by virtue of their recognition of their complement on the pre-existing polymer. This is exactly what a DNA double helix is. One helix is a DNA polymer, and the other helix is its complementary DNA polymer, and they are thus held together by the strong hydrogen bonds between the complement nucleotides. Thus, we have the following:
The complementary base-pairing that occurs between purine-pyrimidine pairs in nucleotides allows for the storage of hereditary information in DNA, since one polymer always contains the information specifying the polymerization of its complement.
This is arguably the most important statement in all of biology, since it underlies absolutely everything in modern evolutionary and cellular biology. Today, it is common knowledge, but imagine how ecstatic Watson and Crick were to come to this realization, for they had discovered the secret of life itself!

RNA can serve as a template of polymerization by virtue of exactly the same mechanism. Recall from above that A,C,T and U are the nucleotides present in RNA. Uracil serves precisely the same function as G does in DNA, and is thus the complement. Although RNA does not serve as the central hereditary molecule of modern biological organisms, it is tremendously important in biology and its importance will become apparent soon.
We have therefore now seen how the chemical structure of DNA, RNA and protein give rise to their important properties. For a protein, this is the 3D structure, and for DNA, is its ability to act as a templated polymer and thus serve as the central hereditary molecule for all life. RNA, as we shall see, has both properties in modern organisms (the ability to form 3D structures which give it particular properties and the ability to serve as a polymer template).
The Structure and Function of Proteins
We have seen how the sequences of amino acids in proteins give rise to their 3D structure. We have yet to discuss how this 3D structure gives rise to the function of proteins. Proteins are the most versatile class of molecules that exist. They constitute 67% of the dry mass of any organism. The 3D structure of a protein is the determining factor of its function. Their versatility is what ensures they carry out virtually all meaningful functions in biology.
Yet before we discuss the vast versatility and nature of these functions, we have to pose a question which will help us comprehend it: What exactly is biological life? If we cannot define this in general, at very least we should be able to provide an Earth-specific definition in terms of the biological systems we know of. We will be able to give a very exact answer to this question later when we discuss the relationship between DNA, RNA and protein, and the concept of biological feedback loops. For now let us work with our knowledge of fundamental processes of templated polymerization to make the following declaration: Biological life is a series of ordered chemical processes occurring within an aqueous media employing a polymeric chemical template which leads to the production of more copies of the template by virtue of templated polymerization. We will stick with this definition for the time being, and it is basically correct. We will modify it later. Note that we have included the proviso about the fact that the fluid medium for all these molecular processes is water.
The continued reproduction of the template requires that a biological system be able to do certain things. First of all, the sensitivity and ordered nature of the reproduction processes naturally requires that the chemical system have a physical boundary which separates itself from the outside world. In modern organisms this function is played by the cell membrane. Note here that we are considering individual cells before overwhelmingly more complex multicellular organisms. In building up our understanding of life from a chemical perspective, we must consider the simplest chemical systems which could be defined as biological first.
This membrane separating a cell from the outside world must allow in certain things from the outside world. The cellular processes that lead to the continued propagation of the template are ordered chemical processes that decrease the entropy of the cell. By the second law of thermodynamics, such processes must invariably increase external entropy of the surroundings. By the principle of energy degradation, which in turn is a consequence of the second law of thermodynamics, the cell can only complete these processes by extracting ordered forms of energy, and converting them into useful work and (by the Kelvin-Planck formulation) rejecting part of that energy back into the outside world which results in a disordering of the overall system (namely, the outside world). The most common energy conversions that occur in the cell are chemical to electrical and vice-versa, and chemical to heat. The general principle we must understand here is that the cell must produce chemical energy in order to perform useful work such as the polymerization of monomers. This chemical energy in turn must come from extracting energy from the outside world. The most obvious way a cell could do this is by employing photons, since they are the most plentiful form of energy. It is no coincidence that the first life forms on this planet where phototrophic. Another obvious way is for biological organisms to harness the chemical energy produced by other biological organisms, but of course this means that such organisms must exist before hand, which is why phototrophes (organisms whose source of energy is light) must exist before organotrophes (organisms like us whose source of energy is other organisms) can.
The reason we have spent so much time discussing this is because it tells us exactly what sort of functions the biological system must perform. Since virtually all of these functions are performed by proteins, it tells us what the proteins in question must be capable above. We are now in a position to talk about this.
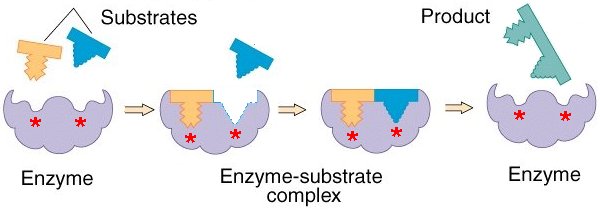
We have said before that in order for the cell to survive as a physical system which runs a cycle of chemical propagation of a polymerized template, sets of ordered chemical reactions must occur in stepwise fashion. The achievement of this depends on the process of catalysis. In general, reactions which occur in biological systems are too slow, if left unaided to occur in the very fast and highly regulated fashion required by sets of highly ordered chemical processes, and many of them are thermodynamically unfavorable. The generation of a new polymer template of DNA is an example of a reaction that could never occur spontaneously were it not for mechanisms which allow life to be thermodynamically possible known as reaction coupling. We will not be discussing reaction coupling here. But we will be discussing catalysis. The maintenance of a system of chemical pathways depends on catalysis for two reasons. Firstly, reactions needed to occur in the space of microseconds would take thousands of years, and second, the providing of an alternate reaction pathway by a catalyst will dictate the pathways taken by chemical substrates in the cell. Thus, the presence and activity of molecules with catalytic properties that act upon certain substrates and produce certain products will determine the reaction pathways which occur in a cell. Such catalysis is the function of a large class of proteins called enzymes. Enzymes are the fastest catalysts known, often speeding up reaction rates by factors of 1014 or more. They are globular proteins where a particular fold in the 3D structure produces an active site whose shape corresponds to the substrate being catalyzed and where the side chains protruding from the pocket induce an unstable transition state in the substrate from which it is more likely to move into the product state.
The relationship between shape and function of enzymes reveals an important principle: Proteins, interacting with each other and with other molecules, form regions which physically fit into the shape of the ligand and which can form a large number of non-covalent interactions with the ligand. A ligand is just a generalized term for a molecule to which a protein binds, since all proteins bind to other molecules.
There are, however, other sorts of proteins besides enzymes. Their varieties are too numerous to cover here. There are two broad structural classes: filamentous and globular. As a general rule of thumb, the latter are those proteins whose usefulness lie in their chemical properties such as their ability to catalyze a particular reaction. The former are those whose usefulness lie in their physical properties such as their ability to provide mechanical rigidity to the membrane. It is important to realize that we are now discussing modern organisms and highly primitive replicating systems during the origin of life would not have these things yet. We are trying to formulate exactly what things a system must have in order to be life and not merely an interesting collection of inorganic molecules. We have seen that the storage of hereditary information by virtue of templated polymerization is one such fundamental property. Nonetheless, in order to discuss biological systems in more advanced terms, we must go into more detail.
We will not be spending a great deal of time discussing structural proteins, as they give a cell support and rigidity and the ability to move. Far more interesting for our purposes of discussing life as ordered pathways of chemical reaction are transmembrane proteins. Transmembrane proteins, as the name suggests, are precisely those proteins which sit across the membrane of a cell. The reason these interest us is because, as we shall see, the maintenance of the pathways of ordered chemical reactions and survival of a cell depend on its ability to integrate and process information from the outside world as well as uptake supplies of energy. This is especially important, as we will see, in multicellular organisms. Structural and transmembrane proteins demonstrate instances where the ubiquity of alpha and beta motifs is useful. Because of their straight, non-globular nature, structural proteins are expected to be comprised of ordered arrays of Beta-sheets, which for the most part, they are. In contrast, transmembrane proteins, having to allow the passage of substance from the outside world, are overwhelmingly based on the alpha helix.
We have seen therefore that biological systems maintain ordered series of chemical reactions in order to survive and propagate. What exactly are these reactions and how do they allow the system to survive and propagate? While there are a tremendous number and variety, there are key points we can take away here. Firstly, all interesting properties of a cell are defined by its macromolecules. It should be clear now that, whereas the DNA holds the information which the cell must propagate by means of its templates, it is the proteins in the cell in any instant which determines the properties it will exhibit. The presence of certain enzymes will result in certain chemical paths being taken, the presence of certain transmembrane proteins will result in certain influxes and responses from the outside world, etc. We are not yet in a position to discuss exactly the nature of the chemical pathways that the cell requires, but we will be soon. For now, the crucial point is that at any instant, the properties of the cell are determined by the proteins present within it. To demonstrate the size and complexity of these macromolecules, shown below is the protein hexokinase (with every atom present in the structural diagram) compared to scale with its substrates adenosine triphosphate and glucose.

There are of course many more classes of proteins. There are motor proteins, which move intracellular cargo in the form of vesicles and are responsible for muscular contraction and a host of other cell locomotion related phenomenon. There are receptor proteins, many of which are classes of transmembrane proteins, which receive and propagate intracellular and extracellular signaling, and, one of the largest groups of proteins, there are DNA-regulatory proteins which bind to sequencing motifs on DNA. All of these are discussed later.
Up until now we have considered the function of DNA and protein independently, as consequences of their macromolecular assembly as linear polymers and, for proteins, their structure. We have seen the key role DNA serves as the central hereditary molecule as a consequence of its ability to polymerize employing a template, and proteins to determine the chemical system constituting the cell by means of maintaining reaction pathways and controlling the relationship between the cell and the outside world. In other words, up until now we have considered nuclei acids and polypeptides independently. There are many more functions of proteins, but we cannot expound on this in any meaningful way, nor can our discussion progress, until we discuss DNA and protein together. Thus, the overwhelming amount of information you are being presented is far from over. But hey, look on the bright side. By now we are past prerequisite knowledge and deep into understanding that has only been acquired in the past 30-40 years. In other words, the third revolution of biology is now what is being detailed.
There is one final note of crucial importance before we move to the relationship between protein, DNA and RNA. Proteins, in general, are extremely specific. A single type of enzyme will catalyze usually only a single highly specific reaction. Different transmembrane proteins pump different substrates across the membrane. Particular motor proteins are specific to certain filamentous intracellular tracks, and many regulatory proteins whose targets are particular other proteins are highly specific to which proteins they target. Without this high specificity associated with proteins, complex biological structures would not be possible. In modern organisms, because their proteins have been so finely tuned by natural selection, they are all highly specific to their particular functions.
The relationship between DNA, RNA and Protein
We have seen that DNA is the central hereditary molecule of modern biological life because of its ability to hold a template to guide its own synthesis. But this by itself is too vague to help answer our question. What exactly do we mean when we say that a nucleic acid contains the information by which biological life is maintained? We know that a string of DNA holds the ability to make another copy of itself, but how exactly is the information which is being copied useful?. The answer is because of the relationship that the nucleic acid sequences of DNA have with the amino acid sequences of polypeptides and the nucleic acid sequences of RNA. To make this task seem less daunting, it may be best to first state exactly what this entails:
1. The ability of DNA to serve as a template not just to guide its own synthesis, but also for the synthesis of RNA
2. The fact that nucleotides can in sequence correspond to amino acids.
3. The ability of RNA to serve as a template for amino acids
4. The ability of RNA to, in a manner analogous to protein, serve particular functions by virtue of their ability to fold into distinct 3D structures.
Proteins give rise to the physical properties of the cell by virtue of their 3D structure which in turn is a consequence of its amino acid sequence. These sequences are very precise to particular proteins and hence particular functions. Thus, the process of creating a functional biological protein is not merely a process of polymerizing amino acids, since the polymer must have a precise sequence in order to be a functional biological polymer.
Perhaps proteins serve as a template for their own polymerization, like DNA? A reasonable first thought, but unfortunately completely wrong. A cursory glance at amino acid side chains tells us that biological amino acids do not possess the same property of complementary hydrogen bond formation that bases do. Proteins cannot serve as templates of their own replication. It must be other molecules which store the information of sequence of amino acids that constitutes a protein. By now it should be clear what this has been leading up to. That molecule is DNA.
Previously it was stressed how tremendously important it was that DNA had the capacity to serve as a template for its own replication. This is still perfectly true. But it is the ability of DNA to serve as a template for the sequence of amino acids in proteins and therefore to serve as not just templates of their own replication but the chemical system allowing them to survive and replicate that makes modern biology possible.
The details of how this is accomplished are based on principles similar to the templated polymerization which is the basis of DNA replication. In other words, the sequence of monomers in a polymer capable of templated polymerization serves to hold the information for the creation of new polymers. We have seen how this templated polymerization can serve to replicate the sequence already present (or rather, its complement) but how does it encode the sequence of a totally different polymer based on a totally different family of monomers? First we shall think about it from a combinatorics perspective. There are only four nucleotides. How is it possible that sequences containing only patterns of four nucleotides can specify sequences containing twenty distinct types of monomers? A single nucleotide cannot represent a single amino acid because there are five amino acids for every nucleotide. Similarly, a sequence of two nucleotides cannot correspond to an amino acid since there are only 16 such distinct sequences. But a sequence of three nucleotides corresponds to 64 distinct possibilities. It is therefore the case that a sequence of nucleotide monomers in a nucleic acid stretch corresponds to a sequence of amino acids by virtue of triplets of nucleotides corresponding to single amino acids. In other words, DNA serves as a substitution cipher. The triplets are known as codons and it is therefore by codons that DNA serves as the template of amino acids.
The process must proceed via the intermediate of RNA, for reasons requiring an understanding of chemical evolution. Of course, RNA monomers possess the same complementary base pairing properties that DNA monomers do (three of the bases are the same anyway. The only different one, uracil, recognizes the same complement as its DNA counterpart thymine).
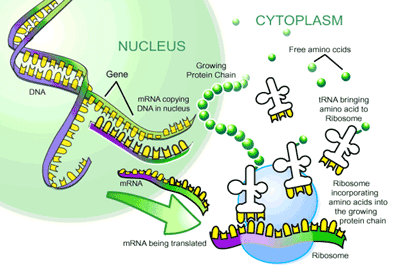
The process by which the ability of DNA to act as a polymer template is exploited to make a sequence of RNA whose codon sequence corresponds to a particular sequence of amino acids is called transcription. The final product of transcription is a sequence of RNA which corresponds to a particular protein is called messenger RNA or mRNA.
The codons of RNA which correspond to particular amino acids are shown below in the table.
GCA GCC GCG GCU | AGA AGG CGA CGG CGU | GAC GAU | AAC AAU | UGC UGU | GAA GAG | CAA CAG | GGA GGC GGG GGU | CAC CAU | AUA AUC AUU | UUA UUG CUA CUC CUG CUU | AAA AAG | AUG | UUC UUU | CCA CCC CCG CCU | AGC AGU UCA UCC UCG UCU | ACA ACC ACG ACU | UGG | UAC UAU | GUA GUC GUG GUU | UAA UAG UGA |
Ala | Arg | Asp | Asn | Cys | Glu | Gln | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val | Stop |
A | R | D | N | C | E | Q | G | H | I | L | K | M | F | P | S | T | W | Y | V |
|
Note that three codons shown above have no corresponding amino acids. Those are stop codons which we discuss later.
The more precise way to summarize all this in the language of computer science is simply that DNA is a 3 bit, base four system.
Thus, a particular sequence of DNA only indirectly codes for a polypeptide. It is the mRNA (which is directly related to its complement sequence of DNA) which directly codes for a polypeptide. Thus, what we see is that a sequence of DNA which is transcribed to form an mRNA which is employed to create a polypeptide is actually the sequence of anticodons corresponding to the mirror image of the mRNA. However, as we shall see, the sequence of codons of DNA, not it's mirror, does correspond to the protein, because there is one more step in the process of protein biosynthesis requiring codon-anticodon pairing in which RNA structures holding the amino acids which are the anticodons of particular mRNA (and therefore, directly correspond to the codons of DNA) assemble on the mRNA.

Thus, consider for example, a sequence of DNA looking like this:
CGACTGCCTGUGAATTTTAGA
Suppose this sequence is transcribed. Then it follows that the mRNA generated will be a mirror of the sequence above and will look like this:
GCUGACGGACACUUAAAAUCU
However, as we shall soon see, it is the mirror of this sequence which corresponds to the amino acid sequence. So, consulting the table above, we see that this generates the following amino acid sequence:
Arg-Asp-Gly-His-Leu-Lys-Ser
Of course, this depends on the fact that we started transcribing at the first nucleotide shown above. This gives rise to the notion of a reading frame. If we started transcribing from the second nucleotide above, we would get the following:
Leu-Thr-Asp-Thr-Stop
We will see later that the transcription process proceeds via an enzyme (actually a family of enzymes) called RNA polymerase, which has the capability to catalyze the formation of the phosphodiester linkage that is the basis of the polymerization of nucleotides. Clearly this means that for proteins to be correct sequences of amino acids resulting in the formulation of useful 3D biological structures instead of useless strings of complete gibberish, the transcription process must be initiated in such a way that the RNA polymerase is positioned exactly correctly, or it will churn out meaningless RNA strings. Such strings are not merely useless, they are dangerous, and by virtue of a series of mechanisms which we do not have the length to discuss here, are destroyed.
It is time now for a very cautionary note. Note that we have barely scratched the surface of the complex mechanisms of transcription. We haven’t articulated how it is started, for one, and how it is stopped and how it is triggered. In fact, we have said very little about it at all. This is true of the bulk of what we are discussing. The notions that are being considered are so complex that we are only elucidating general principles in order to impress upon the reader how fundamental chemical properties give rise to biological ones. This word of caution must be remembered throughout reading this entire piece. Later, the concepts of gene regulation, protein-protein regulation and signaling pathways shall be discussed, but again, only general principles and some examples are considered. It is not possible to go through all the different types of mechanisms and the exact complex interactions that result in them here. Those purposes are served by textbooks whose length exceeds 2000 pages. What I will try to do is cover the principles that underly all types of mechanisms since this is most helpful, but again I must stress-this is not a molecular biology textbook. The reason we are going into molecular detail for processes which are fundamental like protein biosynthesis, allostry and templated polymerization is because without this detail we would be unable to answer the question posed at the beginning. But because this subject is huge, it requires the writer (me) to be highly selective, something the reader should remember.
Anyway, once a correct mRNA string has been formulated, the next polymerization reaction takes place. This is the process of translation, also called protein biosynthesis. Along with DNA replication, this is one of the most fundamental processes that occurs in the cell, and in fact is part of the definition we shall formulate of biological life. The process that follows highlights the crucial importance of RNA.
It has already been explicated that within the cell, RNA molecules are useful for the ability to serve dual purposes as they have the ability of templated polymerization, but also, because the base groupings can form hydrogen bonds with each other (the same complementary bonds which keep DNA double helices coiled) they can form distinct 3D structures. In DNA molecules this never occurs. They always form hydrogen bonds with their complementary helix. They never fold on themselves. This is because that is not part of their function and they are simply too large. As a result they are prevented from doing so because they are virtually always bonded to their complement helice, except when sequences must open so that proteins can operate on them during transcription, replication and regulation, and even then, mechanisms ensure they do not do this, but have do not have time to go through that here.
Because certain RNA molecules have uses not merely as intermediates between DNA and protein biosynthesis, but also for their 3D structure, there are sequences of DNA whose end product is RNA, not protein. As we shall see, there are numerous different types of RNA besides mRNA, such as tRNA, rRNA, miRNA, siRNA, snoRNA and snRNA. All these types of RNA are the end products of the transcription of RNA genes. We will come back to this shortly.
Translation is the central process of protein biosynthesis. It involves the translation of a mature mRNA in the cytosol by a ribonucleoprotein machine called a ribosome. We haven’t come across ribonucleoprotein before, but they play an extensive role in cellular activity. So extensive in fact that they are listed on the very first page as a crucial chemical property giving rise to life. A ribonucleoprotein is simply a biological structure composed of RNA and protein. A ribosome is such a structure.

The pictures above demostrate an important principle about biology in general. It was said before that individual polypeptides are never found by themselves in biological structures. They are always subunits in quaternary protein structures. Let us extend this principle to a higher-order level. Large scale biological structures that can be seen in an electron microscope are composed of many proteins held together by vast numbers of non covalent interactions. These are the supramolecular structures. Many supramolecular structures are assembled from repeating modules of individual proteins. One such example is the capsid. Another example is actin, which is a long fibrous structure that comprises a major part of the cytoskeleton, whose function is to give the cell mechanical rigidity and movement. Actin polymers are comprised of individual actin subunit proteins which, by means of non covalent interactions with each other, can polymerize. This is a remarkable demonstration of how the same principle of constructing polymers from monomers applies not merely to individual molecules in sequence, but also to actual proteins in forming supramolecular structures. There is, however, one important and obvious difference. The proteins serving to give a cell its properties have these properties by virtue of their sequence of amino acids. In contrast, supramolecular structures which are comprised of repeating protein subunits which polymerize have no sequence, since they are composed of identical repeating subunits.
Returning to the previous point The "adapter" between the extensively different organic families of amino acids and nucleotides is an RNA molecule called the tRNA or the transfer RNA. The transfer RNA can recognize both an amino acid, and the codon associated with it. The RNA is translated into protein by virtue of that a set of three RNAs represent an amino acid as a codon. There are 43 possibilities of RNA codons, or 64 permutations. Yet there are only 20 amino acids, which implies that some tRNA can recognize multiple codons or that some codons can recognize multiple tRNA. In fact, both situations occur. Because of the way genetic information is read, there are three possible reading frames for a sequence. The reading frame is critical since it determines the protein that is made. An mRNA read in the wrong reading frame will translate into a nonsense string of amino acids. Although the bulk of the disparity between amino acids and possible codons is remedied by virtue of that most amino acids can be recognized by multiple codons, in several cases, the opposing situation can occur. A wobble base pair mediates this. In a codon-anticodon pairing, where there are three base pairs, one of them can be a poor fit so long as the other two are stable. This improper base pairing is called wobble pairing. It is similar in stability to standard Watson-Crick bonding, but has fewer hydrogen bonds. The most prominent in the codon-anticodon pairing is the GU pair, although another base pair not normally found in RNA, called inosine can also participate.
tRNA molecules are clover-leaf shaped RNA molecules which have numerous unusual bases including pseudouridine. On one end of the tRNA is an anticodon, a three-RNA sequence that is complementary to the codon on the RNA. It is important to realize that the anticodon does not constitute the "end" of the tRNA molecule in the sense that it is not one of the RNA termini, rather it is structurally on the "bottom" of the tRNA, and the anticodon is roughly in the middle of the sequence. The 5' end of the tRNA recognizes the amino acid associated with it, and in this way, the tRNA serves as the adapter between the mRNA which is used to translate the protein, and the amino acids that constitute the protein.

The ribosome is an extensive ribonucleoprotein complex composed of two massive subunits. The ribosome is a machine which combines the reading of mRNA with the assemblage of tRNA in correct order on the mRNA and the petidyl transferase that links the amino acids on the charged tRNA to form functional polypeptides. As with any structure that performs such fine-tuned functions, the ribosome is large and complex. It is comprised of two "subunits", each subunit consisting of two rRNA and the large one having 34 proteins, the small one with 21. The small subunit has slots for the mRNA, the large one holds the tRNA. There are three binding sites called A, P and E. At the A site of the ribosome, a tRNA first binds to the ribosome, at the P-site, which is the petidyl site, the amino acid of the amino-acyl-tRNA within the P-site links to the nascent polypetide, the tRNA of the amino-acyl tRNA complex of the amino-acid previously added being in the exit site. The peptidyl-transferase induces a conformation change gyration such that the tRNA in the exit site is released from the ribosome. This is all shown in the diagram below.

The cylinders are schematic representations of tRNA.
The RNA components of ribosomes are called rRNA, and the genes from which they are transcribed are rRNA genes. The mutual working of tRNA, mRNA and rRNA, exploiting the ability of RNA to serve as a polymer template and a useful 3D structure, is a testimony to how crucial RNA is to the cell.
Having discussed the central processes of transcription and translation at reasonable length, let us step back and review the fundamental principles as they will help us formulating our understanding of biology in terms of chemistry. First of all, the inability of proteins to serve as a template to guide their own synthesis naturally puts directionality on information flow in biological systems. This is shown in the diagram below.
This diagram shows the central dogma of molecular biology. This principle was first elucidated by Watson in 1958. It states that information encoded in sequences of monomers can only flow from nucleotides to amino acids, not the other way around. As we have seen, this is a consequence of difference of molecular structure and functional groups between the two families of monomers. The code in the diagram, by the way, should be interpreted as follows. "Special" means "only done in a lab. RNA duplication does not, in general, occur in the cell.
Unfortunately, we are not yet in a position to fully grasp the importance of transcription and translation. We must turn our attention back to proteins first. We are now just beginning to scratch the surface of modern discoveries.
Oh, and by the way, to those still reading, congratulations on having just passed the 10,000 work mark upon completing the previous paragraph.
Allostery
The structure of a protein is what allows it to hold other molecules and to, by means of noncovalent interactions, bind with a vast variety of surfaces including DNA, RNA, other proteins and small molecules. Yet proteins also overwhelmingly display another function so crucial that I included it by itself as a fundamental property responsible for life. This is the property of protein control. Thus, like everything else about the protein, is a consequence of its 3D structure which in turn is a consequence of its sequence. What this really refers to about proteins is their ability to switch states on the basis of chemical signaling. If the signal a protein receives is in the form of an activator/inhibitor molecule then this is called allosteric control. Allosteric control is one of three major branches of protein control. The other two are phosphorylation and GTPase regulation. We must understand all three in molecular detail to come closer to a very precise answer of what life is at the chemical level. The reason I say precise is because the terms used previously like “hereditary information” and “chemical pathways” are too vague to be helpful in this context.
Let us begin with the fundamental principle of all types of protein control: A protein can exist in different states. Many proteins have two states, these are active and inactive although in enzyme kinetics we usually talk about the tense state T and the relaxed state R. This sort of binary regulation is very common in enzymes, which can be switched on or off and if deactivated can no longer serve as catalysts. For enzymes, such switching between states is the way in which the protein is regulated. For other proteins such as transmembrane proteins, the ability for the protein to exist in different states is a crucial part of its function. For a protein embedded in the membrane to do something such as pump a chemical substance in or out of the cell, it must be able to move between different conformations. Other proteins such as kinesin and dyenin are motor proteins must be able to move across the filamental tracks inside the cell carrying vesicular cargo. The versatile functions of proteins would be impossible without their ability to change state. When we say state we mean a different 3D structure of the protein, in which it exhibits different properties. The crucial principle behind all forms of protein control is that small changes at local sites in proteins can mechanically propagate throughout the protein and induce changes of structure in other parts of the protein. This sort of alteration is the basis of a vast number of biological phenomenon and reveals that proteins have been very precisely engineered over the course of natural selection. Different protein motifs naturally favor particular mechanical changes that can be induced in the protein by the binding of other molecules. The reason that the overwhelming majority of transmembrane proteins are composed virtually entirely of alpha helices is because the shifting of alpha helices against each other (a property beta sheets don’t exhibit) is possible and allows for a mechanism by which the protein moves molecules across the membrane.
In general, all three types of regulation involve a small alteration at one site which amplifies throughout the protein. The precise physical basis for this highly precise physical alterations in protein structure are discussed later.
The first of these three types of regulation, allosteric transition involves regulation of a protein by virtue of the molecules that bind to it. This is most commonly applied to enzymes. There are two main classes of allostery. The first is homotrophic in which the substrate of the enzyme, and the second is heterotrophic, in which another ligand binds to the protein and consequently regulates it. Certain proteins can exhibit simultaneous heterotrophic and homotrophic regulation. There are two sorts of heterotrophic regulation. In positive regulation or activation a ligand serves to turn an enzyme on and in inhibition it serves to turn it off. There are two principle mechanisms by which this is accomplished. Inhibition can either be competitive or non-competitive. Competitive inhibition simply means that the ligand occupies the binding site of the substrate thus making the protein unable to catalyze the substrate. Non-competitive inhibition requires more precise engineering. It depends on a chemical principle called linkage in which shifts in protein structure induced by the alteration of local side chain orientation by the binding of the ligand amplifies to another region on the protein where the substrate binding site is, which will destroy the precise orientation of the side chains required for the binding. This, of course, to be useful at all, is completely reversible. Once the ligand has been released, the protein relaxes and the binding site is free again. Heterotrophic activation works on an identical principle, in which the protein is initially in a state where it is inactive until the binding of the ligand activates it.

Before discussing the other two primary forms of protein control, we must discuss how crucial allostery is to biological life. We have said before that biological life depends on the maintenance of vast numbers of ordered chemical pathways throughout the cell and that these pathways depend on the activity and existence of particular enzymes within the cell. A set of processes like this is too complex to exist without regulation. Allostery is one of several principle processes by which this maze of pathways is regulated. There are three principles ways in which this is achieved. The first is employing feedback loops, the second is employing feedforward loops and the third is employing small intracellular signaling molecules.
Feedback and feedforward loops occur in biology because, as we have seen, enzymes form highly ordered stepwise chemical pathways in the cell the precise nature of which will be expounded upon later. In general, this requires numerous steps and therefore numerous enzymes and enzymes are very specific to their substrates and reactions. This naturally presents the opportunity for mechanisms of control by which the amount of particular products in the cell is regulated. As we have seen, regulating the activity of the enzymes is the mechanism by which the amount of their product is controlled, since without the enzyme the reaction creating the product in question would usually only occur over the course of thousands of years instead of microseconds. A negative feedback loop is an example of control exploiting this. In an allosteric negative feedback loop, a product of a reaction of a particular enzyme is an allosteric inhibitory ligand blocking the activity of an upstream enzyme. By “upstream” we mean earlier in the stepwise pathway that was required to produce the product. Suppose that enzyme 1 takes a substrate A, catalyzes a reaction forming product B, and passes it to enzyme 2 which catalyzes a reaction converting B into C and then passing it to enzyme 3 which takes substrate C and converts it into D. Now suppose that D is an inhibitory ligand for enzyme 1. As a consequence, as the concentration of D rises, the activity of enzyme 1 drops, and the pathway shuts down. As a consequence, the concentration of D falls and the activity of enzyme 1 rises again and so on. This is an example of a feedback loop. A feedforward loop operates on precisely the opposite principle, in which the product of an earlier reaction influences the activity of a later enzyme in a path.
Both feedback and feedforward mechanisms operate throughout the cell as mechanisms by which hugely complex interlocking sequences of pathways can be autonomously controlled by the cell. However, autonomous mechanisms are insufficient by themselves, as we shall soon see. Intracellular signalling molecules are just as ubiquitous as negative feedback loops for regulating pathways, and we will come to those later.
One example of allostery will suffice. The following example is very common in instructive examples because it is understood in atomic detail. Aspartate Transcarbamylase catalyzes the reaction that begins synthesis of the pyrimidines rings in C, T and U nucleotides. One of the final products of this reaction pathway is Cytosine Triphosphate (CTP, which binds to the enzyme to turn it off whenever CTP is plentiful, a standard negative feedback loop. Aspartate Transcarbamylase is a large complex of six catalytic subunits and six regulatory subunits. The catalytic subunits are present as two trimers, each arranged like an equilateral triangle, which are arrayed so that they face each other, and are held together by three regulatory dimers that bridge them.
The entire molecule is posed to undergo a concentrated Allosteric transition between the two conformations, called T (Tense) and R (Relaxed). The binding of the substrates (carbamoylphosphate and aspartate) to the catalytic trimers drives the enzyme to its active R state (from which the regulatory CTP molecules disassociate). The binding of CTP regulatory molecules to the enzyme’s regulatory dimers causes the enzyme to convert to the inactive T state from which the substrates disassociate. The tug of war between CTP and substrate. Thus, this is identical in principle to any basic negative control system, except that as the enzyme is a multidomain protein, a symmetric molecule with multiple binding sites, the enzyme undergoes co-operative Allosteric transition that will turn it on suddenly as substrate accumulates
Each regulatory subunit has two domains, and the binding of CTP causes the two domains to move relative to each other. They function like a lever rotating the two catalytic trimers pulling them closer together into the T-State. When this occurs, hydrogen bonds form between the opposing catalytic subunits which widen the cleft forming the active site within each subunit, which destroys the binding site for the substrate. Adding large amount of substrate has the opposite effect, favoring the R state by binding to the cleft and stopping the catalytic change initiated by CTP. Conformations between R and T are unstable, so the enzyme mostly clicks between T and R. The concentration of these two species depends fundamentally on the concentration of CTP and substrate relative to each other.

The reason we have gone into atomic detail to describe this is because this essay is supposed to convey to the reader exactly the relationship between chemistry and biology. In a subject as precise as this, there is no room for vague terms or imprecise explanations. We cannot toss about notions like “inhibitor” without explaining what this means from a molecular perspective if we hope to have a firm understanding of this subject.
This understanding of allostery is sufficient to continue but unfortunately there are still several more major classes of protein control mechanisms which we must understand for they are crucial to having a firm understanding of this subject.
The reason what follows is so crucial is because whereas allostery involves transitions between states as a consequence of the presence or absence of particular small molecules, the following mechanisms involve the regulation of proteins by other proteins. This is the most crucial aspect of cellular biology. It is estimated that nearly 1 in 3 proteins in human cells have no other purpose except to regulate other proteins (or DNA). Like allosteric transition, the operation of these proteins on their target proteins serves to induce a mechanical confirmation change resulting in an alteration of function of the target protein. Like allostery, this is a wholly reversible effect. Fortunately, we will not spend as long discussing these two control mechanisms because in mechanical terms they operate on the same principle: The small chemical alterations they make to the protein serve to regulate it by altering its structure. Since we have already detailed exactly how this process occurs in general when discussing allostery, we don’t need to go through it again. The principle is exactly the same. Thus, henceforth we will speak comfortably of notions like “the binding of X induces a conformation change in Y” with the assumption that the reader now not only knows exactly what this means, but also appreciates that such mechanisms arise from purely physical properties of the molecules in question. In fact, we will from now on be referring to all causal relationships between large molecules within biological systems with the understanding that the reader now appreciates they arise from conformation changes which can be induced in proteins and the that this is the fundamental principle behind all regulated pathways in biological systems.
The first of these two massive branches of protein-protein control are the kinase-phosphotase pathways. A kinase is a protein which catalyzes the addition of a phosphate grouping to a threonine, serine or tyrosine amino acid side chain on a protein. Many kinases are so specific that they only catalyze particular single amino acids on particular proteins. Other kinases are highly general and can target whole ranges of proteins. Once a kinase has acted on a target protein in this way, that protein is said to be phosphorylated. Like allostery, phsophorylation is employed to, by means of the same mechanisms as just discussed, activate or deactivate a huge variety of proteins. Virtually every protein in the cell can respond in some way to kinases and indeed, many kinases are regulated by other kinases.
On the flip side of this pathway is the phosphotase pathway. Phosphotases are those enzymes performing the reverse reaction to the kinases. The phosphates thus overturn the phosphorylation performed by kinases. Thus the activity of a kinase-dependant protein in the cell depends on the concentrations of the phosphotases and kinases that regulate it. The ubiquity of this control mechanism is difficult to overestimate. At any given time in a mammalian cell, one protein in three will be phosphorylated.

The final regulatory mechanism we come to involves GTPase regulation. GTPases are just as ubiquitous as kinases. GTP is a molecule whose hydrolysis is a highly thermodynamically favorable process, a fact which is crucial in cells as we shall soon see. Proteins which bind and catalyze the hydrolysis of GTP can therefore serve as ubiquitous cellular regulators. Such proteins will bind very tightly to their GTP substrate and will be catalyze the hydrolysis converting it to GDP. This reaction induces a conformation change in the protein which serves to deactivate it. Once a protein is in the GDP bound state it is not energetically favorable to release it. The only way it can be released is via the binding of a protein called a guanine nucleotide exchange factor which induces a conformational change in the GTPase allowing it to release the GDP. Once in this empty state the GTPase will virtually always pick up another GTP since it is in much higher concentrations in the cell than GDP. Such processes naturally make GTP based regulation necessary for the many unidirectional cyclical processes that occur in biology.

Shown above is a typical GTPase cycle in which a GAP or GTPase activating protein hydrolyzes the bound GTP causing the protein to move to the GDP bound state, and a GEF releaseing the GDP allowing the resetting of the system. Thus, the process of GTPase activation and deactivation is analogous to phosphorylation in that it is regulated by the presence or absence of particular GAP and GEF.
Cellular Feedback Loops and Logic Gates
We have seen that proteins can regulate and be regulated by means of phosphorylation and allosteric control and the binding of GTPase. There are of course, many other mechanisms by which the activity of proteins can be regulated in cells. We haven’t even begun to discuss concepts like intracellular vesicular traffic and sequestering, export signal sequences etc. We will not discuss the mechanisms here as that would take too long. We will discuss gene regulation because it is crucial to our understanding, but first we must consider the following. If a single protein could only alter state and conformation on the basis of single allosteric binding, or the phosphorylation of a single side chain, the opportunities for regulation of proteins would be too limited. The ability of the proteins inside cells to respond to stimulus depends on the concept of integration. In other words, individual proteins can alter states on the basis of input of multiple distinct alterations and therefore form logic gates. The existence of cellular logic gates is crucial for the ability of the chemical system to regulate itself. A simple example will suffice. The following example is by far the most common as an introductory demonstration to this principle. There is a protein called cyclin dependant kinase that has a crucial role in the signaling pathways that are responsible for cell division. We come to the concept of signal pathways later. The point right now is that Cdk, like many other proteins, can compute logical responses to sets of conditions (such as the absence or presence of particular proteins which regulate it).
A Cdk only becomes active as a serine/threonine protein kinase only when it is bound to a second protein called cyclin. The fact that the binding of cyclin is necessary for the protein to switch on illustrates a general principle about control mechanisms. We have seen how the conformational states of proteins can be altered by allostery and by phosphorylation and GTPases, but we have yet to consider that proteins binding to each other to make protein complexes can do the same. The fact that proteins form such complexes provides another level of control. For example, allosteric transitions in the protein by means of the presence or absence of certain ligands could induce a conformational change in a protein making it impossible to bind to another protein and therefore perform certain functions. Proteins bind to each by means of surfaces which match due to their folding and side chain orientation of the respective amino acids. Because proteins work cooperatively, the surface contact non-covalent bonds that hold together proteins have been very finely tuned over the course of natural selection, such that proteins which are supposed to bind to each other have precisely matching surfaces, an important notion we come to later when we discuss combinational control mechanisms.
The binding of cyclin to the kinase is only one of three preconditions to turn the Cdk on. A phosphate has to be added to a specific threonine side chain and a phosphate elsewhere in the protein, a specific tyrosine side chain, must be removed. The Cdk is only switched on when all three conditions are met, and in this way it monitors the activity of a protein kinase, a protein phosphate and a cyclin. This sort of signal integration is but one example of a vast array of different sorts of integrative controls but they all follow identical principles. Certain conditions, such as the presence or absence of ligands or other proteins who can regulate the function of target proteins directly or indirectly, allow proteins to act as logic gates. The cyclin-dependant kinase is an example of an AND gate and is shown below:
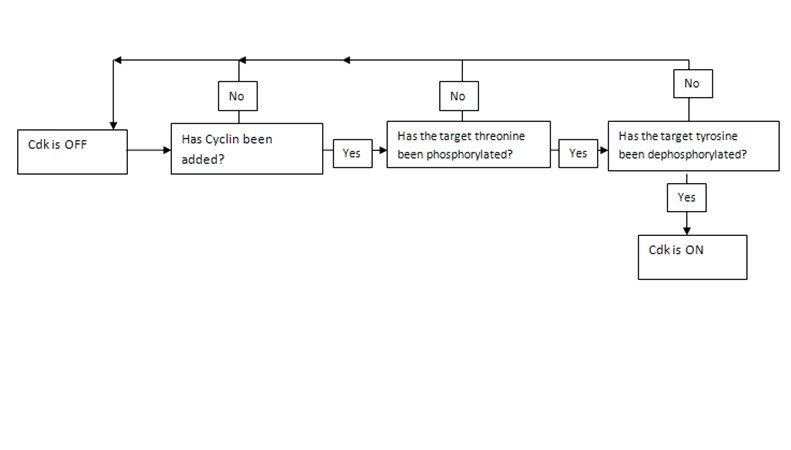
AND gates are very common in the cell. OR gates are rare because they defeat the purpose of signal integration.
Binding Strength and contacts
At this point a short sidetrack into some crucial chemistry is necessary. We have seen that all of these complex interactions between polymeric molecules to allow biological processes to occur depends on non-covalent interactions between the molecules. The ability of enzymes to catalyze substrates depends on the shape of the active site corresponding to the substrate. The ability of proteins to bind to each other depends on their surfaces matching so that side chains can form non covalent interactions holding them together. The ability of proteins to fold also depends on the ability of side chains (and the amide linkages themselves) to form non covalent contacts. In other words, everything interesting that occurs in biology, all of these already discussed complex interactions between large biological molecules depends on non covalent interactions not just holding the correct proteins and ligands together with each other, but ensuring that binding that should not occur does not. The important thing we must understand here is that chemical systems are stochastic. Chemical reactions proceeding depend on the law of large numbers. They depend on molecules colliding in the correct orientation with sufficient activation energy to allow the reaction to complete. Similarly, if we were able to examine systems of large biological molecules interacting with each other on the molecular level, we would see constant collision between molecules within the cell. It would to our eyes look like little more than a confusing hodgepodge of collision. Yet from what we have just been discussing, the cell maintains numerous parallel stepwise chemical processes and complex intricate sequences of chemical reactions.
Given that the cell has to maintain all of these complicated ordered, stepwise pathways in the context of the fact that the cell is a confusing hodgepodge of molecules bashing into each other, how is this ordered existence achieved in a non-orderly cytoplasm? To understand this in any meaningful way requires some understanding of protein kinetics. In all cases where any molecule binds to a protein, it does so by means of a large number of non-covalent interactions with whatever molecule it binds to. This holds true for all interactions. Proteins interact with DNA via non-covalent interactions, with other proteins via non-covalent interactions, with small molecular ligands via non-covalent interactions. The strength of the non-covalent fit between a protein and a ligand is determined by the number of non-covalent interactions, and the precise orientation of the protein for the ligand. If the protein forms a large number of non-covalent interactions, the protein-ligand complex will be more stable than if it was formed by fewer interactions. Thus, protein-ligand interaction is a probabilistic consideration, based on thermodynamics.
This is the primary way in which pathways in the cell are restricted. Proteins and potential ligands are violently colliding all the time in the cell, but only if they actually fit (hence form strong noncovalent contacts) will there be a high probability of the complex being maintained. If not, thermal buffeting in the cell will simply rip it apart. If the molecule doesn’t fit, it is not energetically favorable for a complex to form. As a result, proteins only respond to their specific ligands, be they other proteins, small molecules, or DNA, or (sometimes) all three. Many proteins have active sites which have been so precisely tuned for their ligands by evolution that there is little more that can be done to make the binding stronger.
Thus, noncovalent interactions between biological molecules are the principle mechanism by which distinct chemical pathways are maintained.
Polymerization Reactions
This is a short section in which we discuss the central polymerization processes of transcription and translation in terms of the enzymes making them possible. We have seen how the processes depend on the linking of monomers in linear sequence and that the actual sequence of monomers is determined by templated polymerization in the case of replication of DNA and transcription, and the use of substitution cipher recognition in translation. But there is a general principle underling all processes we should discuss here: The process of creating large linear macromolecules from monomers to serve functions in the cell is a fundamental yet thermodynamically unfavorable process which can therefore only occur in the context of a disordering of the external surroundings. As a consequence, it requires chemical energy provided by metabolic processes such that the thermodynamically favorable conversion of chemical to heat energy drives the thermodynamically unfavorable processes of polymerization. We have already discussed this. We have also seen that the condensation reactions which are responsible for this polymerization are not merely unfavorable, they would be very slow even when made favorable by coupled to a thermodynamically favorable process like the conversion of chemical energy into heat energy. This is a fundamental distinction we must make. Thermodynamic favorability is a fundamentally different thing to reaction rate, a fact that often confounds student being introduced to thermodynamic chemistry. An enzyme can speed up a slow reaction but it cannot make a thermodynamically unfavorable process favorable, since all it does is lower the activation energy. Thus, we now state a general principle governing the process of polymerization in the cell:
The enzyme complexes which are responsible for the linking of monomers in sequence to create biological polymers must be capable of simultaneously lowering the activation energy of two reactions, the first reaction being the actual condensation reaction of polymerization and the second being the release of chemical energy which drives the thermodynamic possibility of the polymerization.
Chemical Pathways
Unfortunately, we are far from finished with the information and knowledge that must be piled on before we can formulate a complete and satisfactory definition of biology in terms of chemistry. Before moving on, it is time to take yet another step back and ask yet again what we have learned and what stage we are up to in defining biological life in chemical terms. We are up to the stage now where we are intimately familiar with the relationship between the central hereditary molecules allowing for their own synthesis, and the macromolecules which actually perform the activities within a biological system that make this replicative process possible. We have also seen that these latter macromolecules exist as part of a vast regulatory network in which they regulate each other and respond to external stimuli, maintaining the interior conditions within the system and producing the stepwise chemical reaction pathways making the replication of biological life possible. In order for long strings of nucleic acid to continue to propagate by means of templated polymerization, they must encode the physical system that makes this possible, in other words, a distinct chemical system with a defined physical boundary in which sets of large and complex macromolecules themselves the product of information within these hereditary molecules, operate. This chemical system is a maze of pathways which allow for the growth, survival and of course reproduction of the entire system by virtue of the information contained within the central hereditary molecules. The maze of pathways is under the control of both external stimuli, internal feedback loops and the hereditary molecules themselves, since they are ultimately what code for the proteins present in the cell which in turn at any given time determine the physical state the cell will exist in. Additionally, the proteins can regulate each other giving rise to yet another level of control. Yet in order for DNA to maintain control over the maze of complex pathways in a cell, it must be able to respond to external feedback from the cell itself. In other words it must be regulated. This is where we hence turn to the crucial notion of gene regulation.
Before discussing the concept of gene regulation, it is time to yet again discard another vague term that has been used up until now. Previously, we have been discussing the idea of “chemical pathways” with the understanding that the proteins present in cells determine these pathways and that they are responsible for the maintenance of self replicating chemical systems. This is too vague to continue to be meaningful. Obviously we cannot detail every chemical process that occurs in the cell. The objective here is to understand precisely what sort of pathways are fundamental to life, so as to improve on our definitions before continuing.
So then, what exactly do we mean by these “chemical pathways”? We cannot cover everything that occurs in modern cells, but we can try to pin down which major types of pathways are fundamental to biology. First of all, we shall remind ourselves what we mean by a chemical pathway in rigorous terms. A chemical pathway is simply a series of stepwise chemical processes that begin with a particular substrate and end with a particular product. This process is determined and regulated by the presence of enzymes acting on the substrates in question to produce the products in question in which we begin with a substrate which undergoes a reaction catalyzed by a particular enzyme, producing a product which is the substrate of another enzyme which catalyzes the reaction of that substrate to make a product which in turn is the substrate of another enzyme and so on. Since these reactions would only occur over thousands of years without the presence of enzymes, the existence of particular substrates in the cell is determined by the active enzymes present. These enzymes in turn are operating in a set of mutual feedback loops with the substrates themselves in order to keep the processes autonomously regulated.
By now it should be clear that biology is defined by the interactions between large scale polymers which have properties allowing for the existence of chemical systems which allows for its own replication. Biology is not simply a process by which DNA replicates. The existence of modern organisms depends on the replication of billions of base pairs over and over again. This sort of process requires the continuous conversion of raw material into chemical energy, because the processes by which the cell creates internal order in a universe tending toward disorder requires the external disordering of the surroundings. Therefore we come to the first of the three major branch of chemical pathways. These are the metabolic pathways. The pathways by which a set chemical system with defined physical boundaries takes up raw materials and converts them into useful energy in order to drive replicative processes and monomers which serve as the materials for polymers which are the products of replicative processes. In modern biology, the central metabolic pathways are the conversion of glucose into NADH, NADPH and ATP.
The useful chemical energy stored in the molecules which result from the metabolic pathways is needed to drive the thermodynamically unfavorable processes that create order within the cell. Since the replication of the molecules whose properties allow for them to hold the template to their own replication is a vastly thermodynamically unfavorable process, these self-replicating molecules must thus serve not just as the template of their own replication, but also for the sequence of other families of macromolecules whose physical properties allow for the maintenance of the set of metabolic pathways allowing the survival and replication of the DNA within the boundaries of the distinct chemical system separated by the cell membrane. Therefore, we come to the second major arm of the three major types of chemical pathways that occur in the cell: The polymerization processes. The polymerization process is necessarily linked to the metabolic process, since the metabolic processes occur due to the existence of the macromolecules produced by the polymerization processes, and the polymerization processes in turn, are made thermodynamically favorable by the products of the metabolic processes. The three major polymerization processes, of course, are transcription, replication and translation.
The third major branch of the chemical pathways that occur in the cell are therefore the regulatory processes. For the cell to uptake raw materials and convert them into ordered macromolecules and useful chemical energy in order to survive and grow requires tight control over the maze of the pathways discussed above within the cell. This requires the metabolic and polymeric pathways to be able to respond to several sorts of control. The first is autonomous control, which has already been discussed, in which metabolic products are regulated on the basis of their presence or absence. Such self regulating feedback loops ensure, for example, that necessary products do not drop to dangerously low (or high) levels. The ability of proteins to respond to such autonomous loops due to the allostery is the first level three major branches of regulation of the cellular processes. The second is internal self-regulation and the third is external regulation. Internal regulation takes the form of gene regulation which we discuss later. The third, which we discuss even later, is of principle importance in multicellular organisms.
Signaling Pathways
We have seen how, by virtue of phosphorylation/dephosphorylation and GTPase based pathways (and other mechanisms yet to be discussed) that proteins can regulate each other. The fact that proteins can regulate each other gives rise to the notion of signal pathways in which activation of proteins “upstream” triggers a set of causal processes leading to some alteration in a process downstream. We have seen how the ability of proteins to alter conformation on the basis of a set of particular chemical condition allows cellular regulatory processes to function employing sets of very powerful logic gates. Now we can extend this understanding to bridge the logic gates together in order that sequences of alterations of conformation of proteins induce causal changes in stepwise fashion to produce some end result. We are therefore leading closer and closer to understanding biology in chemical terms.
We have not discussed the concept of signal pathways yet, but we will since they are crucial to answering the question posed at the very beginning, if you can remember that far back. First we must turn our attention to gene regulation, since signal pathways would not make sense without it.
Gene Regulation
-A genome is much more than a protein code. Within the genome are sequences that dictate where, when and at what rate each gene is to be expressed. Immensely complex pathways of gene regulation are the principle methods by which cells adjust quickly to their external environs.
We have seen that the central polymers which allow the guidance of their own synthesis are substitution ciphers which allow for the creation of polymers whose physical properties allow the maintenance of a chemical system which allows the self replication processes to occur. The other way to state this is that the chemical system supporting the existence and replication of biological polymers which can guide their own synthesis is itself the expression of the substitution encoding within those polymers. Note that I have stated this quite precisely. In some texts it is described as “the proteome (the complement of proteins in a cell) is the physical expression of the genome). I am very hesitant to say something like this because it is very vague and abstract unless read correctly, if read correctly it means exactly the same thing. It is for that same reason that I have been very hesitant to use words like “information”. Instead, I stress the physical properties of the different classes of monomers as the way in which the processes of templated polymerization and the creation of 3D structures with particular chemical properties can occur. Likewise, when discussing concepts like logic gates and signal integration, it is important avoid using vague ideas and stress that these properties are due to mechanical shifts in the physical structure of proteins as a consequence of the binding of other molecules and the resulting alteration of their function.
Here it is time to introduce another piece of terminology. We have seen that sequences of DNA correspond to the sequences of amino acids occurring in proteins. Thus, a genome of an organism (which is its completed gene sequence) contains within it all the sequences of nucleotides corresponding to the proteins present in the organism. A particular sequence corresponding to the code for a single protein is called a gene. (The definition of a gene is actually more complex but this definition serves us well for our purposes here). It is also important to stress that in general, only a tiny fraction of the sequences of nucleotides that constitute the genome of a particular organism are actually direct representations of amino acid sequences in proteins. We do not have the time to go through the details here, but vast strings of sequences of the genome code for nothing at all. Indeed, overwhelming more of the human genome is composed of transposable elements which have been inserted virally and provide a useful record of evolutionary processes.
The fact that genes are transcribed and then translated to form functional biological proteins gives rise to the notion of gene expression. We have seen that proteins can regulate each other by a variety of mechanisms but what should be clear now is that one of the central mechanisms of internal regulation within a biological system is regulation of gene expression. The process by which a gene is expressed to create a functional protein is a multi-step processes which hence has numerous opportunities for regulation. It is impossible for us to go through all of these mechanisms now since that would require us to go through concepts such as DNA compaction, the differences between Eukaryota and Prokaryota, protein folding regulation etc. It is impossible to this in the space of this essay. Instead I will try to elucidate the general principles that underlie the regulation of gene expression by proteins in order that it may help us to define biology in chemical terms.
We have seen that enzymes can operate in allosteric feedback loops with their substrates to give rise to a fundamental regulatory process controlling the presence of particular products within the cell. It makes perfect sense that this notion of a feedback loop can be extended to the relationship between proteins and genes. We have seen in our section on polymerization that it involves the action upon DNA sequences by RNA polymerase which possesses the properties allowing them to catalyze the process of transcription. As a general principle we must understand that DNA is constantly being acted upon by proteins in order that it can be transcribed and replicated (there is another central process called compaction but we cannot go through that here as it will take too long). Thus, gene regulation is the process by which regulatory proteins regulate the processes in which other proteins act upon DNA to transcribe and replicate it. Although the regulation of the action of proteins on DNA by other proteins is a vast subject, like protein-protein regulation it follows general principles which we must elucidate if we are to have any hope of comprehending this subject.
Since our purpose is discussing life in chemical terms, we need to pick the process which is the most fundamental and necessary to understand. That process is transcriptional regulation. Transcriptional regulation works on chemical principles which are in many ways identical to those governing protein-protein regulation. I will state these first and then they will be discussed:
1. Amino acid side chains in proteins can form non covalent bond contacts with the monomers in DNA. This allows for particular proteins to bind to DNA sequences
2. Because A, T, C and G can form distinct hydrogen bond contacts, and because sequences of amino acid side chains form distinct surfaces which can form particular non covalent contacts with particular chemical groups there are sequences of DNA which correspond to binding sites on particular proteins (this has no relationship whatsoever to the fact that sequences of DNA code for sequences of amino acid. They are totally different phenomenon. This is not a form of encoding but is instead a phenomenon where surfaces of particular chemical groups match and form large number of non-covalent bonds because they complement each other). This gives rise to regulatory motifs which are sequences of DNA which correspond to the binding sites of particular proteins therefore allowing those proteins to act on those particular seqences of DNA. This is precisely the mechanism by which the RNA polymerase recognizes where it is supposed to start transcription.
3. Like protein-protein interaction, and the interaction of enzymes with their substrates, the strength of DNA-protein binding is judged by the number of non-covalent contacts formed. If there are too few, it indicates a surface mismatch and no bonding takes place. It is by this that specific target proteins recognize sequences they are meant to act upon.
We are now in a position to discuss the mechanisms by which proteins regulate the expression of genes. Such proteins are termed gene regulatory proteins henceforth referred to as GRP. The number of proteins which are dedicated purely to the regulation of genes is truly vast, even larger than the kinase-phosphotase regulatory class. We know that the amount and type of proteins present in the cell at any given time will determine its particular state. The end purpose of gene regulation, regardless of which step along the path is being regulated, is to control the type and amount of particular proteins present in the cell. Of course, by now we should be merging the different aspects of our understanding. Since proteins can regulate other proteins, and proteins can regulate genes, the result is a set of vast feedback loops in which proteins can control the presence of themselves and each other directly and indirectly by regulating each other.
I have already said that there are numerous steps between the initiation of transcription and the completion of a functional protein and all these can in principle be regulated. However, for most proteins, the most important of facet of gene regulation is regulation if transcriptional initiation. Since transcription produces the mRNA necessary to form functional biological proteins
Of the gene regulatory proteins which serve to regulate transcriptional initiation (which constitute the bulk of gene regulatory proteins) there are two classes, the activators and repressors. Analogous to inhibition and activation in allostery, proteins which are activators serve to facilitate gene transcription.
The simplest of these mechanisms to explain is called a genetic switch. There is a textbook genetic switch which serves to demonstrate the ingenuity of genetic regulation. This sort of genetic switch is part of a large class which serve to regulate transcriptional initiation by binding directly to a site on the DNA where the RNA Polymerase initiates transcription. Depending on the situation, this can serve to block or to facilitate the initiation of transcription. All sequences of DNA corresponding to amino acid sequences are preceded by a region of DNA called the promoter which the sequence to RNA polymerase binds, by virtue of the mechanisms discussed above. The promoter region of particular genes can thus contain other binding sites for the admission of other gene regulatory proteins to bind. The simplest repressors simply physically block RNA polymerase from binding thus preventing transcription. A very simple example to explain is a genetic switch that controls a set of five enzymes that manufacture the amino acid tryptophan. The five genes are arranged to be transcribed as a single mRNA, as a single unit called an operon. A Gene Regulatory protein acts a repressor that switches the gene off, by binding to a sequence in the promoter region called the operator. This acts to directly block the RNA Polymerase, hence stopping transcription. The trp operon is regulated in an ingenious fashion. The GRP does not, under normal conditions, bind to the operator. However, when tryptophan binds to the GRP, it induces a conformational change in the GRP that allows it to bind to the operator, hence blocking transcription. The trp repressor is hence regulated by the very product it regulates, which produces a feedback loop allowing levels of tryptophan to be properly regulated.
The reader should therefore begin to draw parallels between regulation of proteins directly by each other and ligands, and the process of gene regulation. What was shown above is an example of a feedback loop which works on a principle very much identical to an allosteric feedback loop of an enzyme. We have seen that the regulation of proteins directly by each other and ligands is tremendously extended by signal integration. We should expect similar processes to occur during gene regulations. Note that I have repeatedly drawn a careful distinct between proteins being “directly” regulated by each other and gene regulation. The example of signal integration of cyclin binding by Cdk is an example of direct control. Whereas, if a protein serves to regulate the gene coding for another protein, this would be indirect control. Both processes are of course the regulation of the proteins which are present in a cell, since that is fundamentally the purpose of regulation, since the proteins present and active in a cell will determine the properties which it exhibits.
We do indeed find that gene regulation can incorporate signal integration, for without it, complex gene regulation processes would be impossible. However, gene regulation exhibits a particular property which direct protein control does not.
A simple example of signal integration will suffice, one which is also very commonly used for instructional purposes because of its simplicity. Repressors and Activators, as we have seen with the trp operon, can be themselves regulated. Ligand binding can serve to turn them on or off, as can phosphorylation, or the binding to other GRP. Certain operators can be the target of both activators and repressors which hence compete to determine the transcription of a gene. The most well studied example of this occurs in the lac operon, which transcribes the mRNA for a set of genes that control lactose metabolism. But the lac operon is only transcribed when lactose needs to be used as an alternate energy source. The lac operon is under the control of two gene regulatory proteins, one is a repressor, and the other is an activator. Two conditions need to be met for the operon to be transcribed. If the gene was transcribed while glucose was present, it would be wasteful. In addition, if the gene operon was transcribed when lactose was absent, it would again be wasteful to transcribe the gene. So, the two conditions that are required for transcription are the presence of lactose and the absence of glucose.
The operon is under the control of two proteins. The Catabolite Activator Protein (CAP) binds to the operator and activates the gene, whereas another GRP called Lacl, the lac repressor, binds to it under normal circumstances. These two GRP are themselves under the control of two ligands. CAP is normally unable to bind to the operator unless it is bound to two molecules of cAMP (cyclic adenosine monophosphate). The CAP is positively regulated by cAMP and is hence also referred to as the CBP (cAMP binding protein). The lac repressor can bind to the lac operator under normal circumstances. However, when lactose is presented, allolactose binds to the lac operon and induces a conformational change which shuts down the lac repressor. This allows the cell to integrate two signals into controlling the transcription of the lac operon based on two signals and hence allows the cell to compute a logical response to the environs.
This is a tremendously important realization. Not only can proteins act as integrators of signals, so can genes. There are of course far more complex examples of signal integration of gene regulation, but expounding on this more precisely requires an appreciation of the differences between Eukaryota and prokaryota, which we haven’t discussed because our object is to formulate a generalized definition of biological life in chemical terms.
The type of genetic signal integration shown above is competitive signal integration in which genes display different responses depending on which gene regulatory proteins are present or absent. A perhaps even more important form of integration is combinatorial control.
Many times, two gene regulatory proteins with only weak affinity for DNA per se can bind to each other, hence inducing a conformational change which presents a DNA-binding motif to DNA in the proper fashion. This is combinatorial control. Combinatorial control offers another whole dimension into the vast array of possible gene regulatory circuits that can command the transcription of genes based on the integration of a vast number of signals. Multiple individual GRP can form complexes which can activate or repress genes after binding. This allows for the integration of signals from numerous individual proteins which themselves are controlled by certain ligands which have a relationship to the gene being activated (as seen with the trp operon).
Combinatorial control depends therefore on proteins called coactivators or conversely, corepressors. One of the most ingenious facets of of combinatorial control is the fact that many coactivators can bind to different complexes of gene regulatory proteins which serve to control different genes, hence the introduction of particular coactivators or corepressors allows a signal to be sent throughout the genome. Note that coactivators and corepressors are, like activators and repressors, termed as “gene regulatory proteins” since that’s what they do.
The second equally ingenious facet of combinatorial control is that the same coactivator or corepressor can have different effects depending on the proteins which it is complexed with. Thus, a signal affecting multiple genes which is initiated by the activation of repressors or activators by coactivators or repressors can switch on certain genes and simultaneously switch off others, in other words, activators and repressors of gene sequences are themselves activated or inhibited by other proteins that can form part of a combinatorial control complex. This sort of mechanism allows a single protein to induce a major change in the gene expression pattern of a cell.
There is another mechanism, which we do not have time to elucidate fully here, although needless to say it operates on the same general principles already discussed. This mechanism sets gene regulation apart from the signal integration processes of direct protein control. By and large, the latter are binary, in that they serve to turn proteins on or off, but in addition to acting as switches gene regulatory proteins can work on sliding scales in which activator and repressor complexes influence the probability of transcription and therefore the quantity of mRNA transcribed and therefore the quantity of a particular protein present in the cell. Thus we get the picture of a genome of an organism being constantly acted upon by intra-regulated regulatory networks of proteins, the sum result of which is a gene expression pattern which is merely the sum result of the regulation of each gene by the regulatory networks. As we have seen before, the gene expression pattern of a cell will determine its properties. In most cells in most cases, the expression pattern is highly stable, with major alterations occurring, for example, during processes such as replication, in which the cell must double its protein content in preparation to divide. This “sliding scale” mechanism, combined with the process of combinatorial control, gives essentially limitless possibilities for complex regulatory pathways to alter the expression pattern of the cell, and this fact is of critical importance during evolutionary processes.
Having a reasonable appreciation of gene regulation now, it is time to return to our discussion of signaling pathways. We previously discussed signaling pathways in terms of proteins inducing conformational changes in each other in causal chains by virtue of mechanisms previously discussed, but we never discussed where those pathways ended. Now we know. The end of a signal pathway is called the target protein. Many signaling pathways have target pathways which are gene regulatory proteins. Thus, some initial signal inducing a conformational change in an initial protein could be relayed to inducing some long term effect by altering the transcription pattern of the cell.
Applying the principles of feedback on multicellular levels
Despite the fact that I have stressed the cut-down and generalized nature of the material presented here, it is nonetheless true that by now the reader’s understanding is relatively sophisticated. There is no reason why we cannot discuss, albeit at a rather rudimentary level, multicellular organisms. Previously, we had more or less been considering individual cells as distinct chemical systems with discrete boundaries. In fact, everything we have talked about thus far pertains to one cell. Multicellular organisms are constructed of many cells which must be coordinated in order to produce such complex actions as the lifting of an arm, or the creation of a memory. Everything we said before is completely general and therefore applies to the individual cells present in multicellular organisms but clearly there must be another entire level of control in order to make complex multicellar organisms possible.
Previously, we have considered the internal and autonomous regulatory processes that occur within a cell that result from the interactions of proteins which each other, with gene sequences and with ligands in which the state of a cell, defined by the activity and presence of proteins within it, responds to internal stimulus. However, we have yet to make any discussion of external stimulus. This is still important in organisms which exist as single cells. For example, bacteria must be able to respond to external signals in order to move up the concentration gradients of particular chemical environments, a process called chemotaxis, but clearly the process will be far more crucial in the existence and survival of multicellular organisms.
Despite the overwhelming vastness of this topic, there are some general principles which we seek to elucidate here. First of all, we can fortunately apply knowledge that we already have. Regardless of the nature of the external signal, there is something common to all of them. All external signals will induce an effect on a cell by means of initiating a protein cascade in the target cell. We have already discussed these cascades at length. What we haven’t discussed is that many of these cascades, especially in multicellular organisms are initiated not by internal regulatory processes but by extracellular signals. Fortunately, the manner in which these are initiated by these extracellular signals employs principles which (surprise, surprise) are identical to those we have already discussed, in which extracellular ligands induce conformation change in proteins initiating the protein cascade. There are two primary ways that extracellular signals can do this. The first is by ligand diffusion through the membrane in which the ligand molecules diffuse through the boundary of the cell and bind and activate target proteins within the cell. We have seen that many of these proteins initiate cascades which eventually end up targeting particular gene regulatory proteins which end up regulating particular genes (which probabily end up regulating other genes or proteins. If you haven’t caught on by now, regulation is rather important in cellular processes), One such example of this is the action of hormones in multicellular organisms which travel through the bloodstream and diffuse into target cells where they activate particular nuclear hormone receptors.

The second major mechanism for extracellular signalling is by employing transmembrane proteins. We haven’t discussed transmembrane proteins much because we haven’t considered exterior conditions very much but they will come up now. The transmembrane proteins we are considering are the vast class of proteins called signal transducers. As we have come to expect by now, though signal transduction is a huge topic, there are general principles we can elucidate. All signal transduction by transmembrane protein works on the fact that changes on the part of the protein which faces the outside can induce changes on the part of the protein facing in the inside of the cell. The latter alterations can then activate protein cascades.
One example will suffice. There is a ligand called alpha interferon which can activate a pathway called the Jak-STAT pathway. There are actually many Jak-STAT pathways which are activated by different ligands which are part of a class of signal ligands called cytokines but we shall take this one as an example since they all operate on the same principle and because this path was first discovered via the action of interferon. The Jaks are called so because they are Janus kinases after the two faced Roman God (for appropriate reasons as we will see in a moment) and STATs are Signal transducers and activators of transcription. Again appropriate. Let us see how it works. We have seen that proteins are in general composed of multiple polypeptide chains (cast your mind back to protein folding and quaternary structures). A protein composed of two polypeptide chains is a dimer.
As we can see from the diagram below, the dimers of the signal transduing protein below are initially separate. This protein belongs to the class of cytokine receptors and is in fact the largest class of a set of signal transducing proteins which are called enzyme-linked receptors. The binding of the ligand induces a conformational change in the protein subunits which allows the two to dimerize, or to come into contact. This allows the Jaks to cross phosphorylate each other as is shown in the diagram. Casting our mind back to protein-protein regulation we recall that many proteins can be activated by phosphorylation including those which themselves are phosphorylators of target proteins. That is what this process does. The active Jaks can now phosphorylate their target which is the receptor they are bound to which provides a binding site for the STATs. If you cast your mind back to protein-protein regulation again, recall that in signal integration we discussed that the phosphorylation of proteins could alter their ability to bind to other proteins since the site could either induce a conformation change in the protein or provide a site for the other protein to dock.
The rest is shown in the diagram. Note how this process is quite typical in that we begin with an extracellular ligand inducing a conformation change in a transmembrane protein and the signal being relayed to alter a pattern of gene expression.
Signal Integration
Signal integration has come up time and time again and we should not be surprised that it comes up again here. We will not spend long on this since the general principles we have already been through are identical. In general the ability of proteins to act as signal integrators means that not only can they function as integrators of intracellular signals, but extracellular ones. Previously, in our example with Cdk, the signals which the kinase integrated were both from the inside. But of course we know that extracellular signals can induce protein cascades as we saw above with Jak-STAT, and that protein cascades end with particular target proteins. It makes perfect sense therefore that proteins which integrate the signals of extracellular ligands do so by being the target protein of multiple parallel cascades simultaneously. The principle is identical to Cdk being switched on by the three conditions that must be met, except that in this case, the conditions that a protein will integrate must be initiated from the outside to trigger the cascades that end with the conditions being met.
Cell differentiation and signaling types
We are nearly at the end now. To those still reading, congratulations on having passed the 20,000 word mark.
All of the general principles discussed thus far pertain to all cells. However, multicellular organisms are composed of different types of cells with different phenotypes. What exactly do we mean by different types of cells? By now it should be very clear. It has been stressed time and time again that a cell is determined entirely by the proteins present within it, which is why that particular thing is in the end what is being controlled by the vast regulatory pathways present in the cell. Thus different cell types are determined by different gene expression patterns. This is in fact the solution to a puzzle that had confounded biologists for some time. During the embryonic development of a multicellular organism, the central process is that of differentiation in which cells develop into their distinct phenotypes. Biologists initially thought that during the developmental process, cells undergoing differentiation selectively lost sequences of genes so as to gain distinct transcription patterns, but this is wrong. It was experimentally verified that all cells within a multicellular organism regardless of their phenotypic differentiation contain the same genome. Thus the process of cellular differentiation depends on the alteration of gene expression patterns so as to create unique complements of proteins in particular cell types. In most cell types, approximately 20,000 of the 30,000 proteins in the human genome are expressed. The particular proteins which are expressed, and their levels of expression, thus determine the cell type under discussion. Previously, we have discussed how the gene regulatory pathways create distinct gene expression patterns but what was perhaps not stressed enough is that many of these patterns are permanent, and not just permanent to the cell being differentiated, but also to the cells created once that cell duplicates. In this way, particular gene expression patterns can be preserved through generations, despite the fact that the initial extracellular signal which caused them occurred a long time, even years ago. This is the fundamental concept of cell memory. Thus, the general principle we must understand is that the permanent process of cellular differentiation occurs because initial transient extracellular signals can induce permanent alterations in the gene expression patterns of particular cells which can then be passed onto progeny.
The mechanisms by which cells do remember expression patterns are too numerous and complex to elucidate fully here, but like everything else in molecular biology, they do obey the general principles we have already talked about. A very simple example will suffice here. Suppose a particular protein serves as a gene regulatory protein to regulate its own synthesis in a positive feedback loop. In other words, suppose for example that the promoter region of this gene cannot allow the RNA polymerase to initiate transcription without the presence of the gene regulatory protein which is being coded for by the very sequence under discussion. Thus, under normal circumstances, this protein will not be part of the gene expression pattern, since it requires itself to initiate its own transcription. But once a transient extracellular signal allows the transcription of this protein to proceed, it will continue to maintain its own expression long after that signal has dissipated. Not only that, it will maintain its own expression in the daughter cells of the initial cells once that cell has duplicated because the proteins present are inherited during duplication. This rudimentary example of cell memory is a ubiquitous mechanism by which cells differentiate.
This rudimentary example however does not by itself do justice to the ingenuity of differentiation, because cell differentiation incorporates the processes of signal transduction and combinatorial control in a very clever way. Recall that combinations of gene regulatory proteins could serve to regulate the expression patterns of certain genes. Thus, the introduction of gene regulatory proteins to some cells but not others is a principle mechanism by which cells are differentiated by virtue of position. Let us suppose we have two clusters of cells A and B which are initially identical. Now suppose that cluster A is in a region of high concentration of a particular gene regulatory protein and cluster B is not, such that the GRP is inducted into the cells of cluster A and is responsible for the transcription of particular genes which will not be present in cluster B. Those genes in turn could code for gene regulatory proteins which in turn could, by means of combinatorial control, serve to give rise to distinct expression patterns that cluster A will possess and cluster B will not. Of course we could keep going and suppose that some of the gene regulatory proteins coded for by the genes activated by the proteins which were initially activated by the initially inducted GRP are themselves gene regulatory proteins and so on. We are witnessing the power of combinatorial control to propagate expression patterns because combinatorial control allows for individual gene regulatory proteins to propagate large scale alterations in the cell’s expression pattern. This is not just hypothetical. This is in fact the mechanism by which an entire organ (the eye) is formed.

The above diagram shows how clever this mechanism is. If we begin with distinct cell clusters only one of which has a particular GRP introduced, then within one generation we will have two distinct expression patterns resulting from the induction of the GRP, and we have seen above that this GRP, by means of combinatorial control, could by itself introduce a major alteration in the expression pattern of cells into which it is inducted. Now suppose that a new GRP is introduced into the daughter cells of both distinct generations of cells that arose after the initial induction of GRP 1. In this case it is clear that we will have four distinct expression patterns, and continuing this argument, with another GRP introduced to the daughter cells of all four cells of generation 3 will result in eight distinct expression patterns, all because the initial clusters differed by the introduction of a single GRP! This demonstrates the power of the combinatorial control process, and is a reason that combinatorial control has been and still is critical in the evolution of multicellular organisms. I use "still is" because evolution is of course an ongoing process.
Conclusion
We finally come to the end, the most important part of this essay. Because we are now in a position to answer properly the question posed at the beginning! There is no more information to absorb. The information at this point is hopefully all in your head, it just needs to be organized into a complete set of underlying principles. You, the reader are no doubt exhausted, but try to appreciate the enormity of what we have just done! We began with nothing more than a few fundamental electrical properties of atoms and just now we were discussing how the eye forms! That I was able to elucidate this entirely is testimony to the vast explanatory power of reductionist science. The fact that all of what was spelled out above is now so deeply ingrained into the modern scientific understanding that I was able to write the entire essay off the top of my head is testimony to the rigor that was put into making these discoveries.
Although a huge amount of information has been presented the reader should note that while there was much content, the underlying principles behind everything we discussed have not changed. Molecular biology is a very logical subject. The same underlying principles occur over and over and over again. For example, having just checked using the Word Find tool, I have counted some variant of the word “polymer” appeared over 400 times in this essay. Some form of the word “regulation” appeared nearly 300 times. Thus presumably these concepts are rather important. The point is, in the course of this we came back to the same principles over and over and over and over again. The fact that proteins can alter their conformation on the basis of the binding of other ligands and the binding and action of other proteins is a fact which has come up in everything we discussed. The relationship between polymers capable of guiding their own synthesis and replication and polymers whose 3D structure allow for a chemical system making this replication possible has also come up countless times. As we moved to ideas as complex as cell differentiation, the underlying principles remained identical. We were still discussing proteins inducing conformational changes in each other and binding to other polymers by means of non covalent contact. Not much of a surprise. These chemical principles come up again and again because they are the fundamental chemical principles defining biological life and are thus the answer to our question. The complexity of molecular biology lies not in the underlying principles but in the combinatorics of the processes.
Thus we can now state in exact chemical terms what biological life is. We have in fact already stated it with varying degrees of precision, but now we shall be able to formulate it exactly. Let us first return to the six chemical properties I listed at the very beginning:
1. The ability of nucleic acids to act as a template for its own synthesis by polymerization and the synthesis of related nucleic acids by virtue of complementary hydrogen bonding and hence the ability of DNA to act as a template for RNA
2. The ability of nucleic to act as templates for the synthesis of proteins by virtue of triplet sequences of nucleic acids which correspond to particular amino acids
3. The ability of proteins to, by means of non-covalent interactions between carboxyl, amine and side chain groupings, to fold into distinct 3D shapes giving them the totality of their individual properties
4. The ability of proteins to form hydrogen bonded contacts with the outer face nucleotides on DNA polymers and hence for the protein molecules to alter the activity of regions of DNA (the contact of which can induce allosteric changes in the binding proteins)
5. The ability of RNA molecules to form ribonucleoprotein complexes by virtue of hydrogen bonded contacts with matching surfaces of polypeptides
6. The ability of polypeptides to form protein complexes by means of matching surfaces via hydrogen bonding and hence for allosteric mechanical changes to be induced in the binding proteins or small ligands resulting in a change of structure and function,
This should no longer be opaque and meaningless. This should now make perfect sense to the reader. These are chemical properties resulting from the orientation and polarities of certain atoms in molecules which are what underlie all of the vast number of processes which we have just discussed. This has been stressed repeatedly throughout the essay. I said at the very beginning that the single principle underlying all six of these was the ability of matching surfaces of covalently bonded polymers (which exist due to their complementary 3D structures which in turn are consequences of the sequence of chemical groups present) to form large scale non covalent contacts. This fact should now be very clear to the reader since we discussed processes like allostery, signalling, catalysis and translation in atomic detail stressing this very fact.
So, this list of six constitutes the chemical principles which give rise to biological systems. Thus we can now define a “biological system” in chemical terms. Some of this will have already been presented above, but the point is that it is instructive that everything is now incorporated now that our understanding is sufficient to appreciate all of it.
Let’s begin in general terms. I think it’s reasonably fair to say that you’ve had enough of the stream of text. So let’s start our final definition of biological life with a nice diagram:
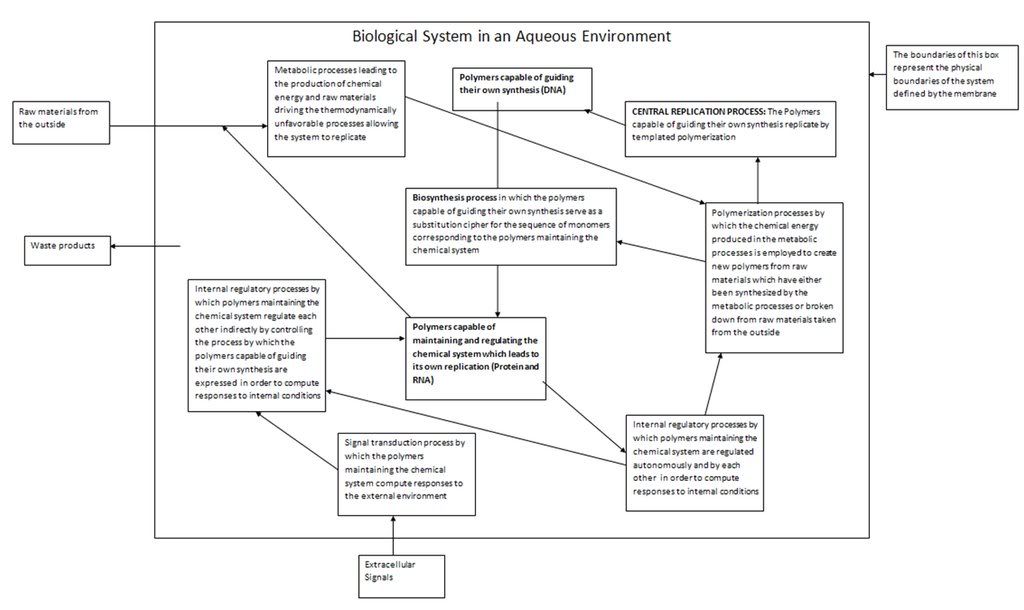
It is important to stress that this diagram is by no means complete. As I have stated before, we have by no means investigated all of the processes occurring in the cell. We have not discussed central pathways such as proteolysis, or given any thought to the huge number of posttranscriptional control mechanisms, we have not considered the differences between Eukaryota and prokaryota, and the fact that the former keep their DNA in a nucleus. We have not considered the fact that there are vesicles which are topologically distinct from the cytosol such as the Golgi apparatus. We have not discussed the endocytosis and biosynthetic-secretory pathways. We have not considered the function of the endoplasmic reticulum. We have not discussed the synthesis of phospholipids to construct the membranes of the vesicles and the cytosol. We have not discussed the cytoskeleton and the functioning of motor proteins in processes such as vesicular transport. We have not discussed the processes by which nuclear export is regulated in Eukaryota. We have not discussed the relationship between differerent types of nuclear RNA in eukaryote. We haven’t discussed the importance of telomerase. We have given no thought to the actual process by which a replisome is constructed and by which chromosomes are duplicated. We have not gone into any discussion of the cellular processes and phases by which the whole cell is replicated. There is a whole level of regulation involving the mechanisms of intracellular protein sorting and signal sequences we have not talked about. We haven’t discussed the central process of intron splicing and its importance in posttranscriptional regulation or its important link to chemical evolution. We haven’t considered the crucial pathways of repair and recombination which occur when strands of DNA break or there is improper copying. We haven’t talked at all about the process of compaction of DNA in Eukaryota involving histone octamers and the formation of chromatin. We haven’t discussed the relationships between activity and compaction of DNA such as in the formation of heterochromatin, or the signal processes involved in the compaction of DNA. We haven’t discussed the processes of homologous duplication and recombination which are so crucial for evolution by natural selection at the molecular level. We haven’t discussed the importance of regulatory combinatorics in evolutionary processes. We have only given a rudimentary consideration of cell differentiation and haven’t considered advanced concepts like imprinting or the formation of Barr bodies. We haven’t actually talked about chemical evolution and the formation of DNA-protein based systems from primitive ribozyme based systems. In other words, there is a lot we haven’t talked about.
Nonetheless, the diagram above does show the fundamental processes defining biological life. What we have spent the last 22,000 word elucidating are the crucial links between the list of six properties above which were first presented at the very beginning, and the processes in the diagram shown above. Let’s try to sum up these 22,000 words. The view of life that has been developed during this discussion is one which is highly computational in nature. Any interesting occurrence in the body can be characterized in terms of interactions between proteins, proteins and DNA, proteins and RNA. That's it. Molecular biology is little more than the study of those three molecules and their interaction, and anything at all in life can be characterized in terms of them. Cells operate by means of a complex set of feedback loops, switches, control mechanisms, signal integration and computation. Each cell is simply a microprocessor. Every cell in molecular biology can be characterized as a complex set of computational commands. Consider the following diagram:
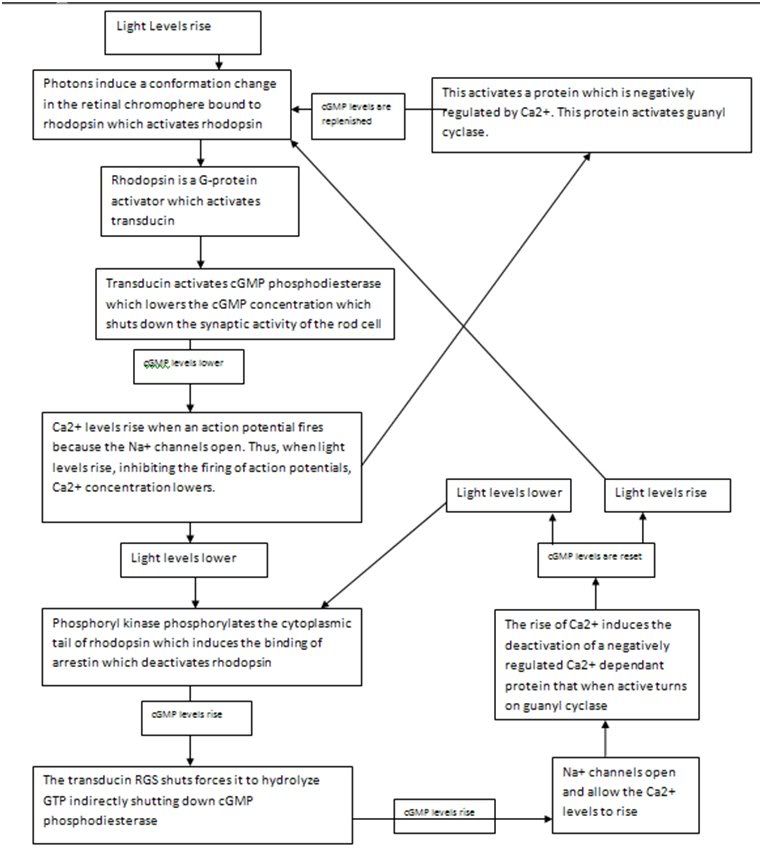
This diagram shows the process by which light from the outside induces the firing of the optic nerve. The important thing to notice is that the whole process is described as a causal chain in which the involved proteins respond to particular signals to produce outputs on the basis of the alterations that the conditions cause in their structure and function. As we have seen, this is absolutely crucial. The ability of proteins to alter their structural conformation on the basis of the conditions of the cellular environment underlies everything interesting in molecular biology. It is for precisely that reason we can say something like "A signal...which induces...which induces...which induces..." etc. These causal chains characterize every single thing occurring in the cell and the body, and they are the object of our study. These sorts of causal chains where proteins activate each other, control DNA which in turn controls the proteins, RNA, etc and vice-versa can be used to explain anything we want to in molecular biology, which is exactly what we have just been doing. Everything which has been discussed has been characterized in terms of complex causal processes, each step depending on a set of conditions, which makes the cell a very powerful microprocessor. In this way, every process in the cell can be characterized essentially as a set of logic gates. Consider the following diagram. You may not know what the particular proteins and genes labelled in it are but you should be able to identify the general processes which occur. We can see ligands entering the cell and binding to target proteins (in fact, nuclear hormone receptors) so as to activate gene regulatory proteins serving to facilitate transcription. You should also note that this is an example of genetic signal integration by combinatorial control and therefore an example of extracellular signal integration since the gene is switched on by the binding of two proteins activated by different pathways.

Thus, what we have elucidated thus far allows us to make the following statement: Biological systems are sets of integrative computational feedback processes of chemical systems controlling and allowing for the growth and replication of chemical systems which have the computational capacity to compute responses to their external and internal environments.
And with that, I shall close with a rather anticlimatic ending:
That, ladies and gentlemen, is biological life.
"Physical reality” isn’t some arbitrary demarcation. It is defined in terms of what we can systematically investigate, directly or not, by means of our senses. It is preposterous to assert that the process of systematic scientific reasoning arbitrarily excludes “non-physical explanations” because the very notion of “non-physical explanation” is contradictory.
-Me
- Printer-friendly version
- Login to post comments